🧬 AP Biology Unit 6
12-16% AP Weighting
Topics
6.1 DNA and RNA Structure
6.2 Replication
6.3 Transcription and RNA Processing
6.4 Translation
6.5 Regulation of Gene Expression
6.6 Gene Expression and Cell Specialization
6.7 Mutations
6.8 Biotechnology
6.1 - DNA and RNA Structure
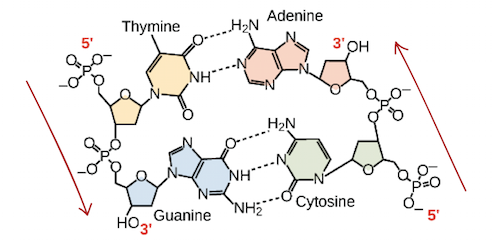
Nucleotides are made of Pentose sugar, Phosphate group, and a nitrogenous base. There are 5 nitrogenous bases, U, T, C, G, and A. U Is only used in RNA, it replaces T in DNA. Phosphodiester bonds connect a nucleotide to another at the 3’ prime. (The Pentose sugar “points” to the 5’ end, it ends at the phosphate group) DNA is double stranded, RNA has one strand.
DNA strands are antiparallel.
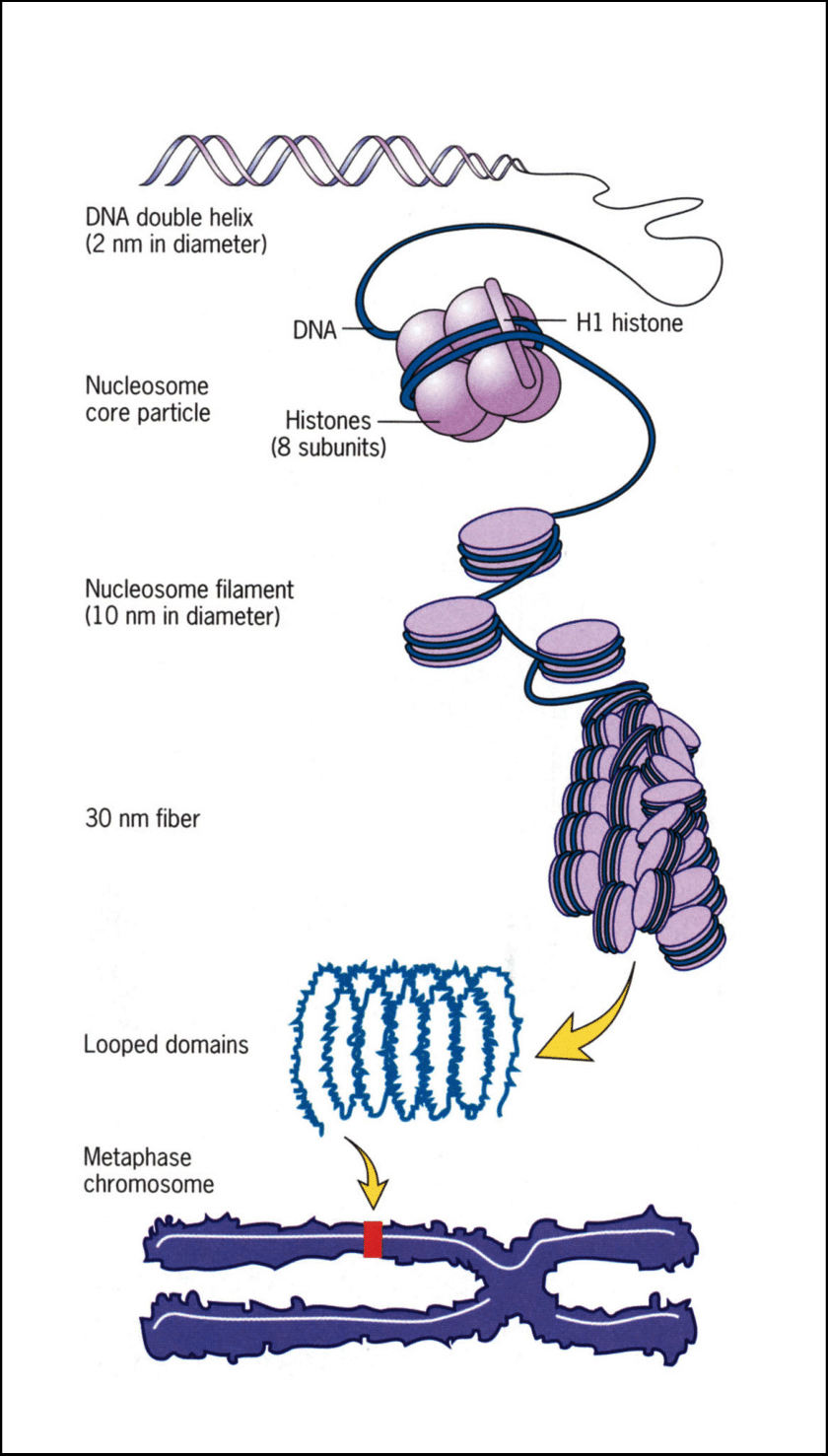
DNA wraps around histones and nucleosomes and twists to form a chromosome. Multiple histones form one nucleosome.
5.2 - Replication
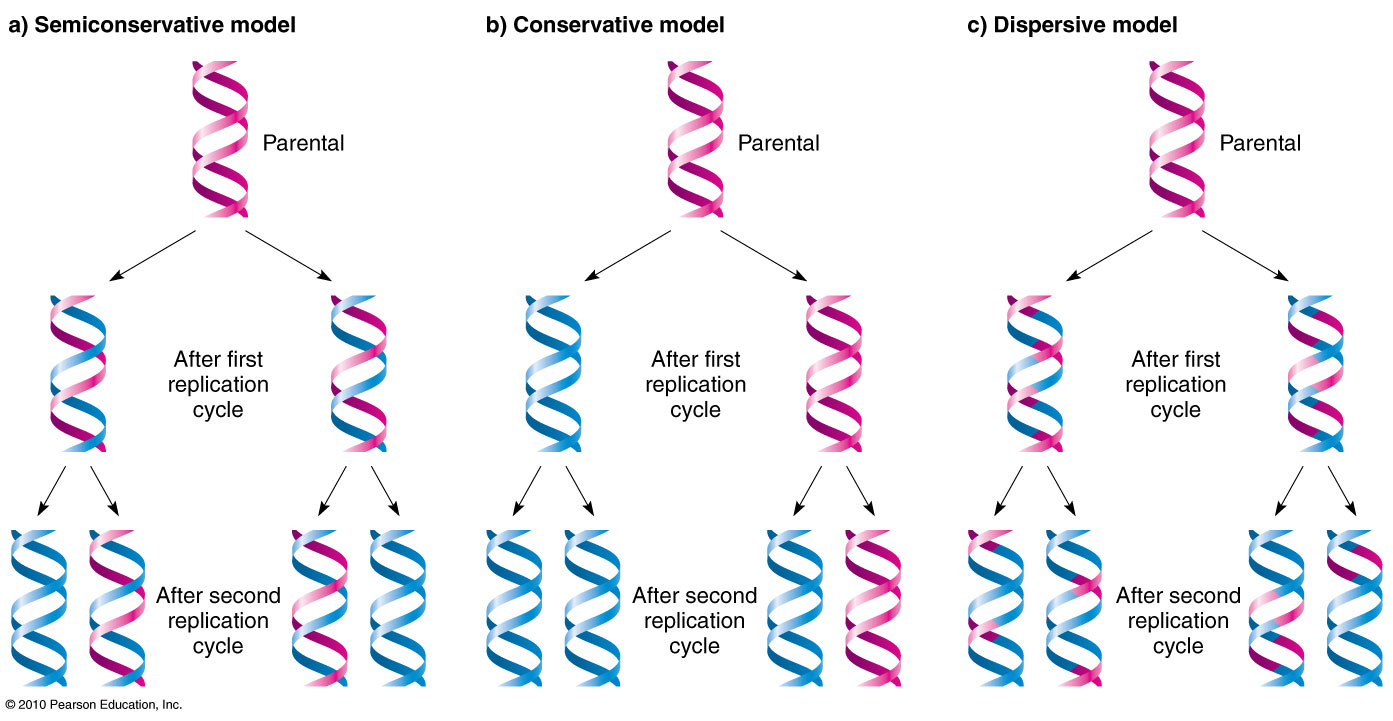
AP Biology follows the semi-conservative model of DNA replication. The original strand is called the parent strand, and it produces the daughter strands. DNA replication actually starts in the middle of the DNA, in what’s called in origin of replication, or ori. A ‘bubble’ is formed, and each end of a bubble is called a replication fork.
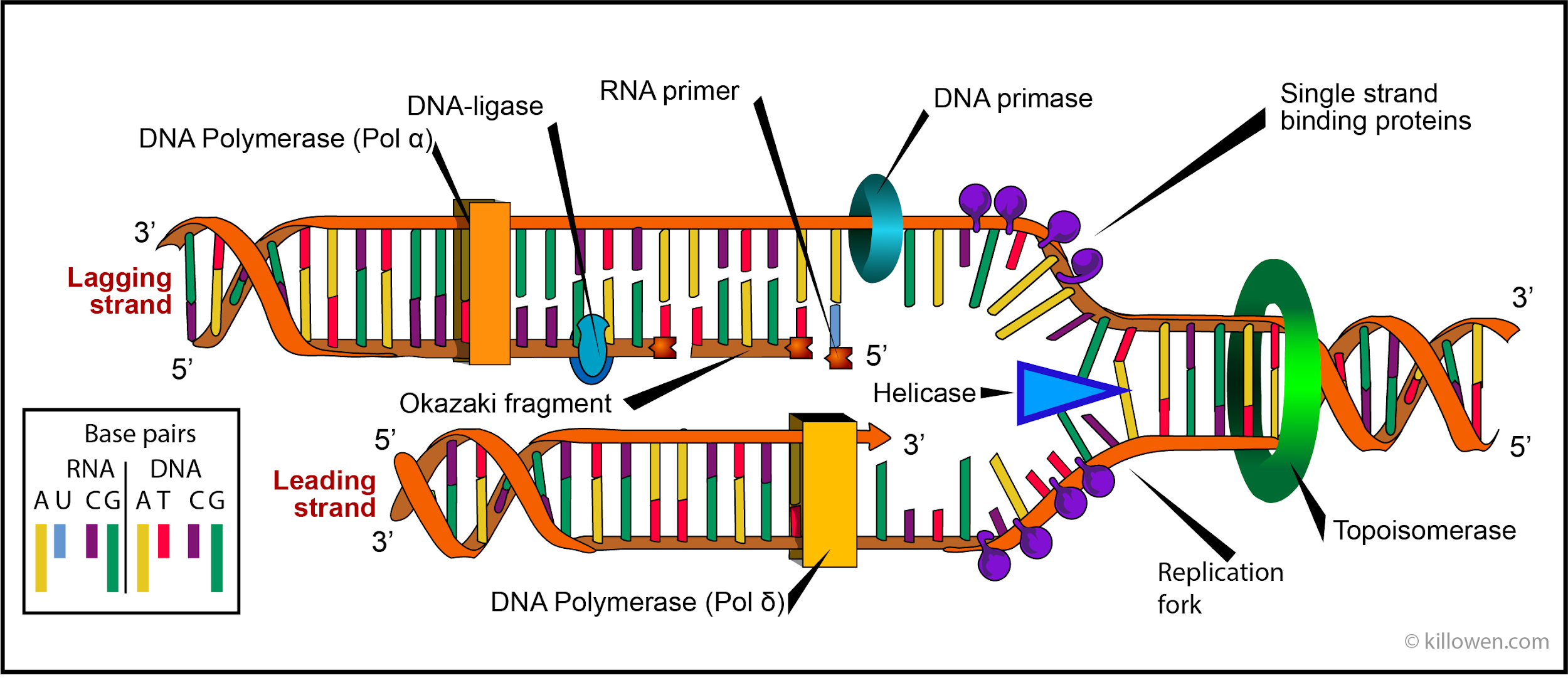
Enzymes involved in DNA replication (in order):
Topoisomerase
‘Unwinds’ or ‘Untwists’ the DNA. The DNA goes from a helix to a ladder shape.
DNA Helicase
Opens the DNA, splitting at the hydrogen bonds between two nitrogenous bases
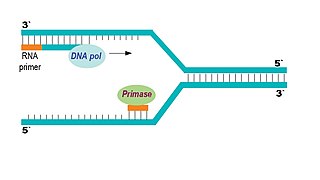
DNA Primase
Creates an RNA primer for the polymerase to bind to
DNA Polymerase
Binds to the RNA primer and forms a new DNA strand, reading the DNA from 3’ to 5’, creating 5’ to 3’. The “leading” strand has one continuous polymerase, while the “lagging” strand has several.
Ligase
Connects Okazaki fragments with Phosphodiesterase bonds.
Nuclease
Looks for errors/mutations in the DNA and removes them. Also breaks down mRNA.
5.3 - Transcription & RNA Processing
Translation is similar to replication, but instead of a second DNA strand being created, a mRNA strand is created. In addition, while there are many proteins involved in replication, nearly all of the jobs of those proteins are done by RNA polymerase in transcription. (Unwinding, opening, transcribing, closing, retweeting DNA
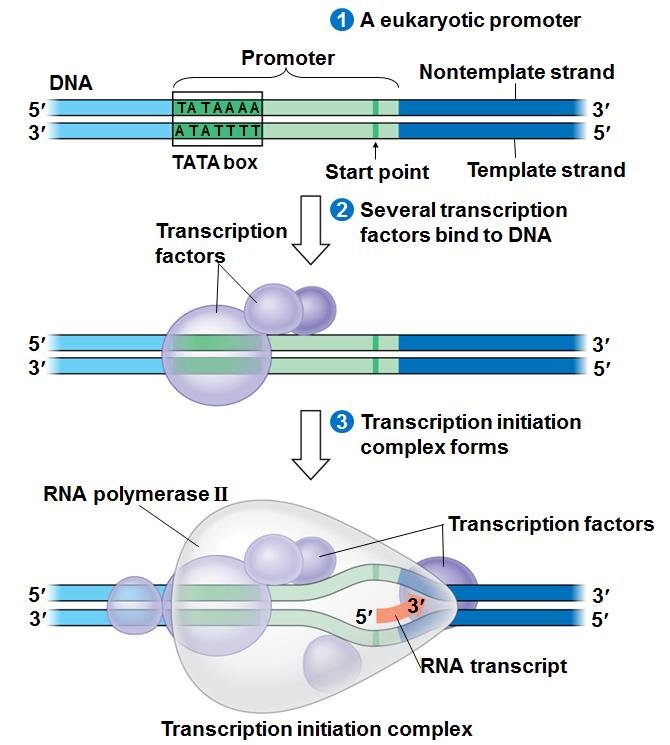
The start of a gene, where RNA polymerase attaches, is called a promoter. It starts with a TATA box. Transcription factors recognize the TATAAAAAAA… sequence of nucleotides of the TATA box and binds to it. The RNA polymerase binds to the transcription factor to form a transcription initiation complex.
The RNA polymerase creates pre-mRNA from the DNA, and does the job of untwisting, opening, replicating, closing, and retwisting the DNA.
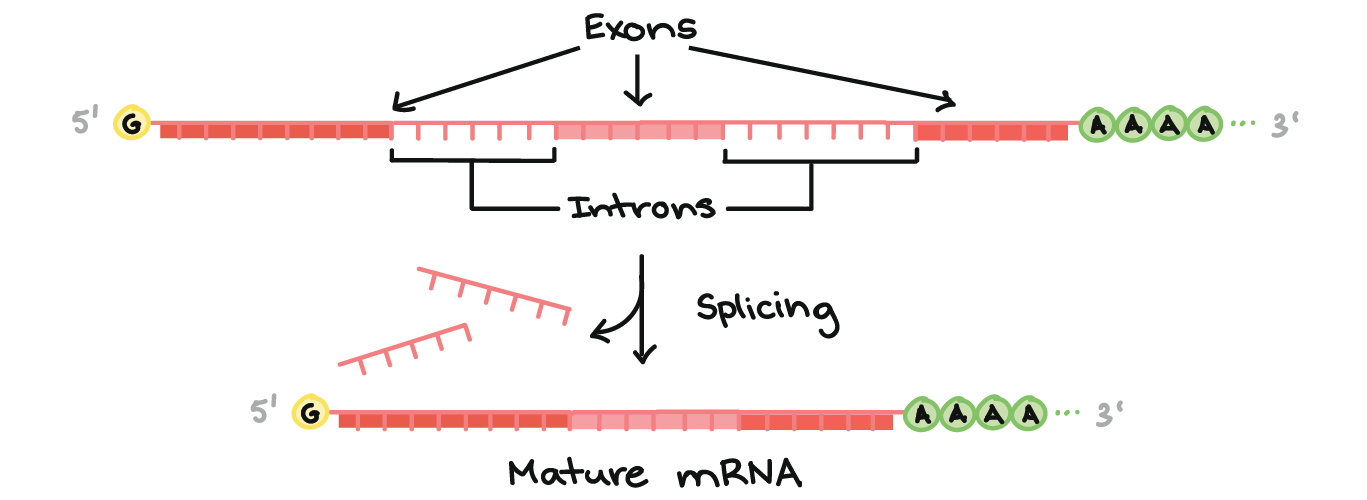
The pre-mRNA is created by transcription. Pre-mRNA has introns, which are DNA segments that do not code for a protein, they are removed by a spliceosome. Then, to prevent the RNA from being degraded too quickly by nucleases, a 5’ (5 Prime) cap of GTP is added to the 5’ end of the mRNA which prevents degradation on that side. The other side has a poly-a tail, which is a long chain of adenines. The adenines will slowly be broken off first, buying the mRNA time to be translated. It is now mature mRNA.
How Transcription is Involved in Virus Reproduction
Viruses rely on the host’s transcription abilities to replicate themselves. Retroviruses are even more complex.
Viruses are in a genomes in a capsid and are not cells. There are different shapes of viruses. Some have a viral membrane, which is a cell membrane that has been stolen from a host cell. Glycoproteins stick out from the viral membrane. Viruses are obligate intracellular parasites. They need to infect cells to reproduce. Prions are infectious proteins. Virions are naked DNA. Bacteriophages, or phages, only attack bacteria. Phages reproduce via the lytic or lysogenic cycle.
Lytic Cycle
Phages will inject their DNA/RNA into bacteria, and the bacteria’s Polymerase will transcribe it, making many viruses. The bacteria breaks down its own DNA/RNA to make more of the virus DNA. Eventually, the viruses break out of the cell, killing it.
Lysogenic cycle
The lysogenic cycle does not kill the host, making it a less serious infection. The virus will inject its DNA/RNA into the cell, and the cell incorporates the virus's DNA/RNA into its own. The virus (DNA/RNA) is now called a prophage. When the cell divides, the virus DNA is in more cells, and when the DNA is transcribed, the cell enters the lytic cycle.
Retroviruses
Retroviruses are the most complex viruses and use RNA. They have reverse transcriptase, which does transcription backwards. This allows them to easily insert themselves into our own DNA. They are now called a provirus. Eventually, this DNA will be activated, killing the cell and making for viruses. HIV is an example of a retrovirus.
6.4 - Translation
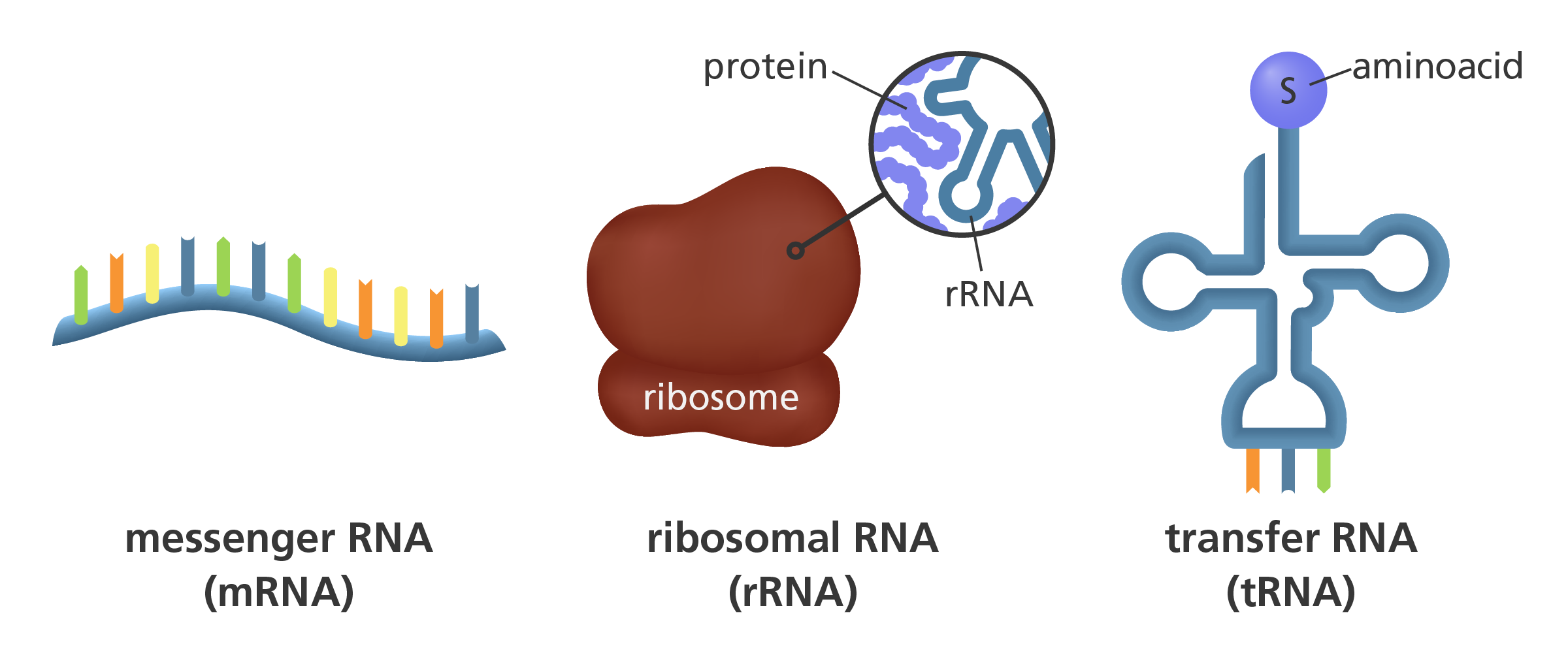
There are three major forms of RNA: mRNA, tRNA, and rRNA. Spliceosomes are made of snRNPs, which are made of snRNA.
snRNA -> snRNPs -> spliceosome
The processed mRNA made by transcription is captured and translated by a ribosome, which is made of rRNA. The ribosome attaches a tRNA codon for each mRNA codon. The tRNA has a base to attach and the corresponding amino acid. The amino acids will split from the tRNA and be a chain of amino acids, a protein. The tRNA can get a new amino acid to continue the process. The amino acid is combined to tRNA with aminoacyl-tRNA synthetase. 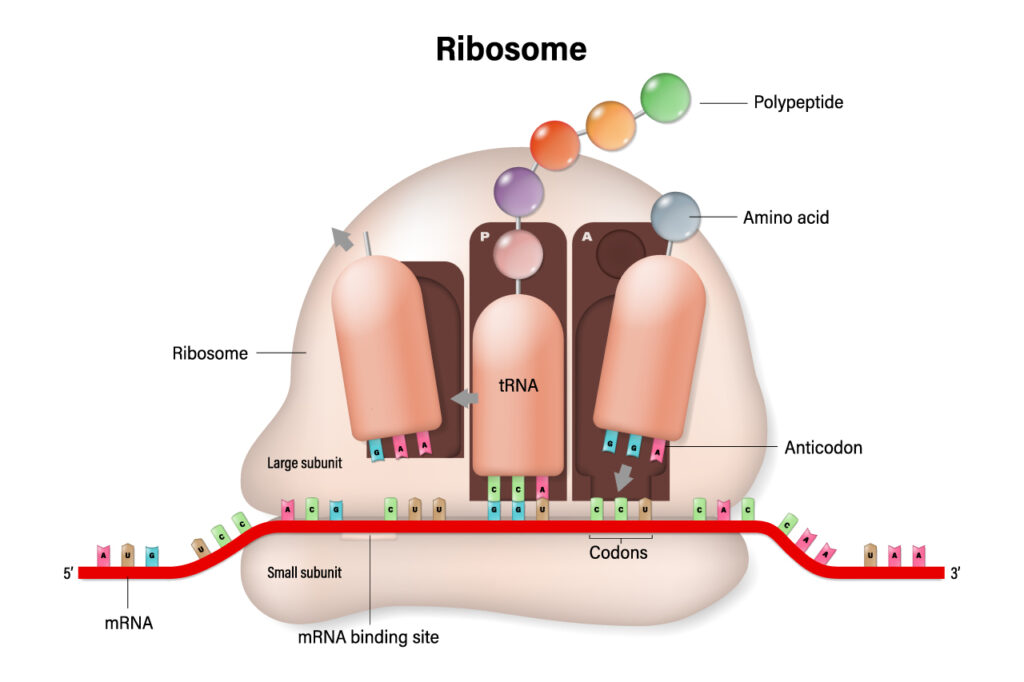
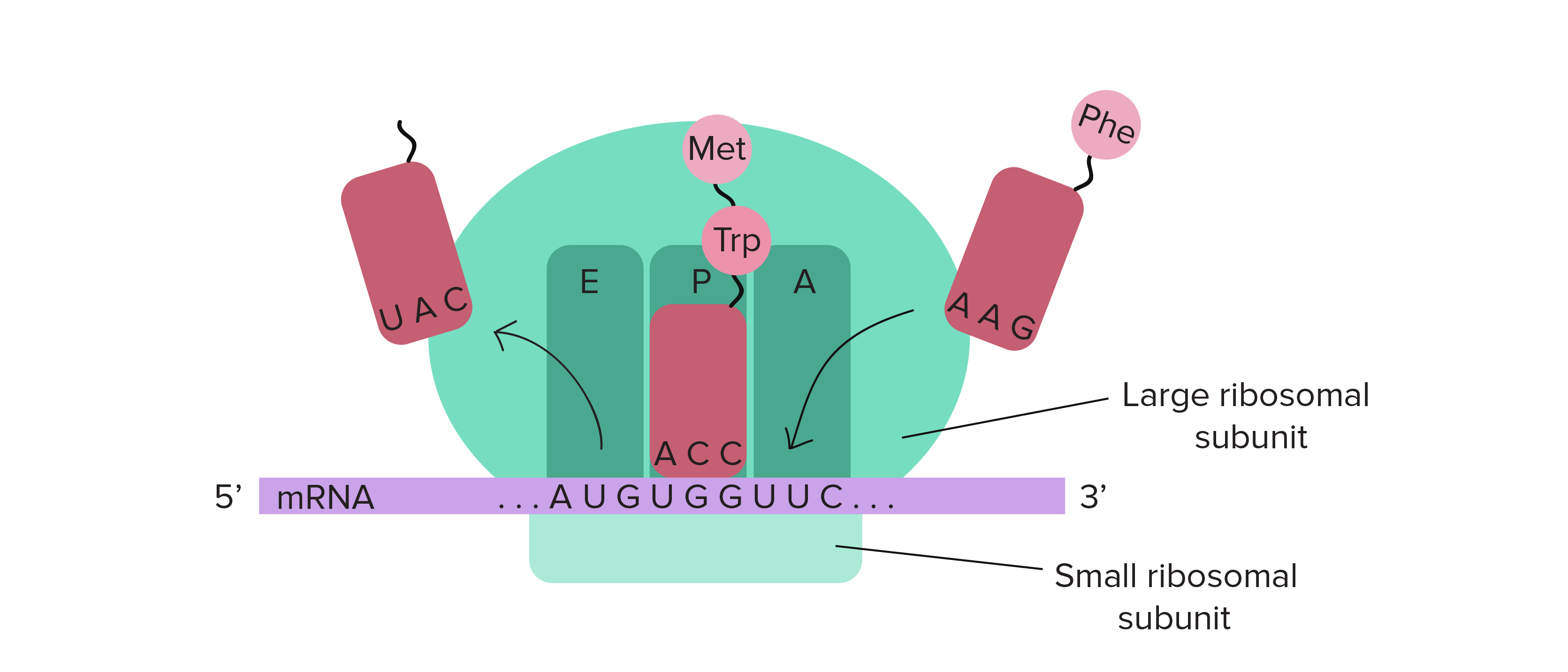 Remember EPA for the ribosome sites. A is where tRNA enters, P is where the peptide bonds forms, and E (Exit) is where the tRNA exits.
Remember EPA for the ribosome sites. A is where tRNA enters, P is where the peptide bonds forms, and E (Exit) is where the tRNA exits.
6.5 - Gene Regulation
Gene Regulation in Bacteria
Gene regulation allows bacteria to modify their metabolism by stopping the production of enzymes. Since metabolic pathways are driven by enzymes. If you stop making the enzymes, there are no more pathways.
Bacteria use operons to regulate their gene expression. An operon is the segment of DNA containing multiple genes and a promoter region. The promoter region is made of the TATA box and operator. The operator works as an on-off switch, when an active repressor (repressor/regulatory protein) binds to it, the genes cannot be transcribed.
Operons are either inducible or repressible.
Inducible Operons are always off and require an inducer to activate them. The Lac operon is an example of an inducible operon. The regulatory gene for the Lac operon is always active and makes repressor proteins that bind to the operator. Lactose binds to the repressor, making it inactive. This allows the genes for lactose digestion enzymes to be transcribed.
Repressible Operons are always on and require a repressor to turn them off. An example of this is the TRP Operon. The repressors for the operon are inactive and need a corepressor, in this case TRP, to make it active and repress the gene. Tryptophan acts as the corepressor and prevents more of itself from being made when there is too much.
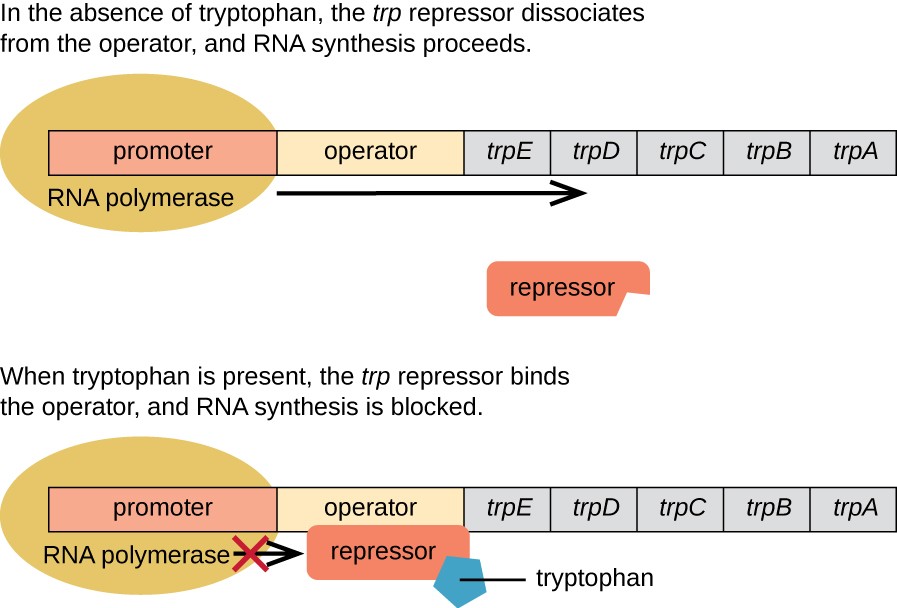
Gene Regulation in Eukaryotes
Eukaryotes rarely have an operon, but gene repression is still similar. An operon is the idea of multiple genes with a single promoter and an operator, but in eukaryotes most genes have their own promoter and no operator. Instead of the repressors binding to an operator, they bind to a silencer, which can be many base pairs away from the promoter, but when bonded, a DNA bending protein will bring the silencer over to the promoter to block transcription. There are also enhancers, similar to silencers, which have activators (transcription factors) that bind to them. Genes with an enhancer are often still translated without an activator, but at levels so low it's not an issue. The enhancer increases the translation.
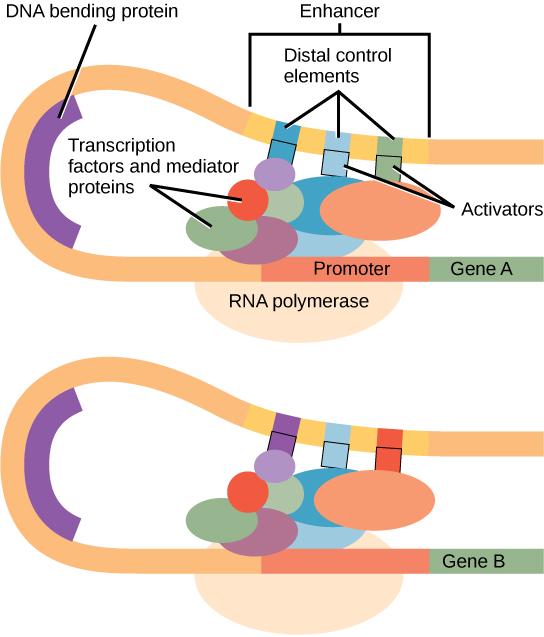
Transcriptional Regulation
Transcriptional regulation involves transcription factors binding to specific DNA sequences to regulate gene expression.
When an activator (transcription factor) binds to it, enhancers will increase transcription. A DNA bending protein will bring the enhancer over to the gene, making it easier for RNA polymerase to bind.
Repressors will bind to Silencers, which decrease transcription by preventing RNA polymerase from easily binding to the promoter.
Eukaryotes also use non-transcription regulation methods for regulating genes.
DNA Methylation
Attaching methyl groups to DNA causes DNA to become too tightly woven together to be easily transcribed, so that segment of DNA is effectively “turned off.”
DNA methylation is actually passed on to children, which is how certain people have a higher chance for developing certain conditions. Some genetic risk factors are the nucleotide sequences, but some are the methylated segments.
Histone Acetylation
DNA is woven around histones. In order to access that DNA for transcription, an acetyl group is added to the histones, loosening the DNA around the histone. This allows the RNA polymerase to access the DNA for transcription.
RNA Processing
RNA processing involves alternative splicing and microRNA regulation.
Alternative Splicing: removing introns and joining exons to produce different mRNA transcripts
RNA interference (RNAi): microRNA is binding to complementary mRNA sequences to block translation or degrade mRNA. RNAi is a process, not a type of RNA.
Homeobox (HOX) genes are an important regulatory gene for embryonic development, they control where body parts develop and how many they develop. If there is a problem, a human may have extra limbs or organs, or they may have limbs in areas they should not. One example is the Sonic the Hedgehog Gene. (This isn’t a joke. It’s the easiest to remember though!)
6.6 - Gene Expression and Specialization
Gene regulation controls whether a gene is expressed. If a gene is translated and transcribed into proteins in sufficient levels, that gene is expressed. Inhibition of translation or transcription as well as the increased production of a different protein to inhibit or destroy the original protein can prevent gene expression.
There are many different types of cells in humans (and most multicellular organisms). The cells in your stomach must have very different gene regulation than the genes in your brain, otherwise your brain would produce stomach acid and destroy itself. However, all cells contain all of that organism's DNA. Specialization is when a cell becomes a specific cell style (ex: red blood cell). Stem cells are unspecialized cells that can divide and then specialize out. A zygote is a stem cell. There are a few types of stem cells:
Totipotent (total)
A totipotent cell is a cell that can develop and become a complete (total) organism, like a human zygote.
Pluripotent
A pluripotent cell can become any type of cell
Multipotent (multiple)
A multipotent cell can become a few (multiple) different types of cell. Bone marrow is an example of this type of cell.
6.7 - Mutations
Types of mutations
Point Mutation - A single nucleotide is changed
Synonymous Mutation - A change in the nucleotide sequence occurs, but the old codon and new codon still code for the same amino acid.
Insertion Mutation - A new nucleotide is inserted into the sequence
Deletion Mutation - A new nucleotide is deleted from the sequence
Loss of function Mutation - A protein can no longer complete its function
Gain of function Mutation - A protein has a new function
Missense Mutation - changes the amino acid a codon codes for
Nonsense Mutation - changes an amino acid codon into a stop codon
Frameshift Mutation - An insertion or deletion causes the rest of the nucleotides in a gene to shift left/right one space, shifting many into different codons. Ex using words: The Fat Cat Ate The Big Hat -> Ath Efa Tca Tat Eth Ebi gha t. It is now gibberish. The spaces can represent the split between codons.
Inversion Mutation - A segment of DNA is somehow flipped/inverted, instead of being ABCDEF it is FEDCBA. This generally happens because a segment gets broken off and then reattached but in a flipped order.
6.8 - Biotechnology
Polymerase Chain Reactions (PCR)
PCR can make many copies of a specific segment of DNA or specific genes. DNA is heated until the hydrogen bonds break and the strands separate. Then, replication occurs. This speeds up replication by not requiring helicase and being able to target just a small segment of DNA instead of replicating everything. The DNA is connected to unnecessary parts because Ligase is not used.
Gel Electrophoresis
Gel Electrophoresis is used to sort DNA by charge or by size. The gel used is porous and allows the DNA fragments (often from PCR) to move through it. An electrical charge is run through the gel, attracting the negatively charged DNA to the opposite side. The smaller the DNA segment, the further it can travel. This allows us to see the sizes of different DNA segments. Genetic profiling, including paternity tests, are done through gel electrophoresis.
DNA Sequencing - The Sanger Method
The Sanger Method was the first method of DNA sequencing. First, PCR creates many copies of the DNA strands. It uses diDNA, which end the DNA sequence as more nucleotides cannot be attached to it. The diDNA comes in different forms of nitrogenous bases like DNA. Because of this, we can add one type of diDNA base to a set of DNA and use gel electrophoresis to determine the base pairs. AT the end of the gel, the diDNA lights up depending on the base, and the smallest are read first because of gel electrophoresis. This is repeated on many replicated DNA strands until we manage to get each base as a diDNA, and therefore know its base.
Conjugation
Conjugation is the “mating” of bacteria to produce for genetic diversity. The bacteria will cut out a small portion of their DNA, replicate it, and form it into a plasmid they can give away. Through the Pili, two bacteria exchange plasmids. A commonly traded gene is the genes for antibiotic resistance.
Restriction Enzymes
Scientists use Restriction Enzymes (nuclease) found in bacteria to cut up DNA. Bacteria use Restriction Enzymes to cut out viral DNA. The restriction sites where the enzymes cut are palindromes.
3’ CATATG 5’
5’ GTATAC 3’
Enzymes are specific to their restriction sites. They can cut DNA straight in a blunt end or staggered in a sticky end. Sticky ends can easily attach to other sticky ends and form new hydrogen bonds.
Cloning
Plasmids are often used as cloning vectors. They are the mechanism to get DNA into another organism. We take DNA from another organism, such as a gene that produces human insulin, turn it into a plasmid, and give it to another organism. The DNA is now recombinant DNA as it comes from multiple organisms. Now, we can make bacteria produce human insulin to help diabetics. The bacteria are now GMOs, recombinant/transgenic bacteria.
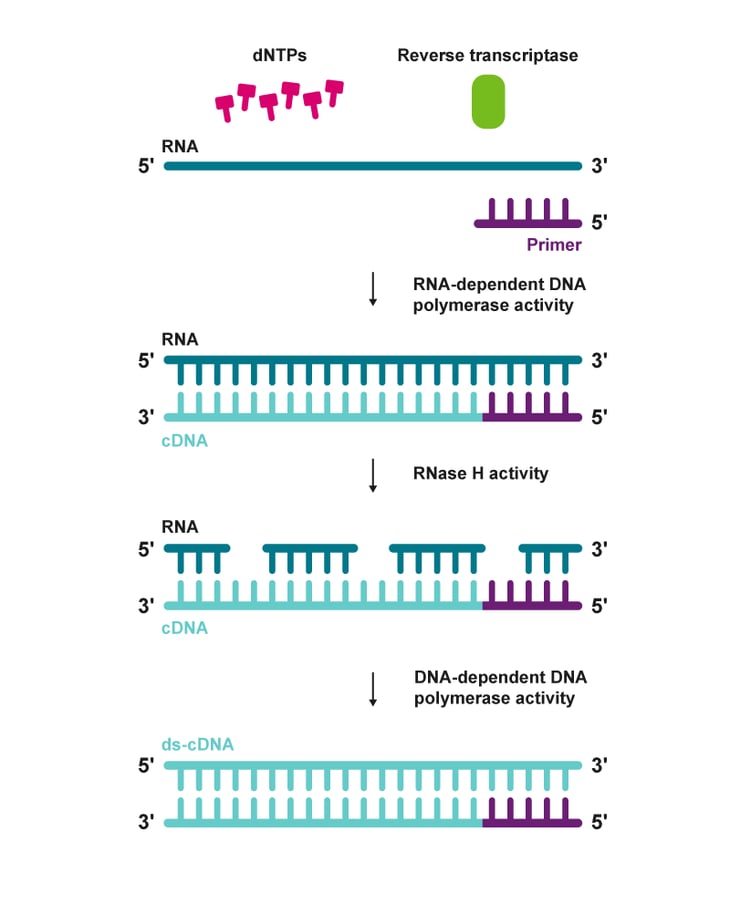
We now use yeast instead of bacteria for some things because bacteria will transcribe the introns, leading to messed up proteins. However, we can get around this with complementary DNA (cDNA). Scientists take mRNA after it has been spliced and use reverse transcriptase to make intron-free cDNA.
Cloned animals
To clone an organism, you can take the DNA from a totipotent (stem) cell (2n) and replace an egg's (n) DNA with it. The egg is given a little shock to simulate the force of a sperm hitting the egg so replication and division begins.
Dolly the cloned sheep had issues and died young because she had “old DNA.” She had arthritis and other diseases associated with elderly sheep.
Cloned animals do not always look and behave the same as their “parent.” For example:
Identical twins have different personalities
Chance the bull was a very friendly bull, but his clone was not
Cloned Calico cats will have different patterns. This is because of the different arrangement of Barr Bodies, which X chromosome is crumpled?
Transgenic animals are animals who have been given DNA from other organisms. They can produce large amounts of otherwise rare substances. These substances, often proteins, get added to the animals milk, and these animals can be milked to get this protein.