biochem 5 amino acids, primary structure, protein, and separation techniques
1/63
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
64 Terms
what are 4 main functions of proteins
catalysis (enzyme)
transport
structure
motion
example of catalysis function in proteins
Enolase (in the glycolytic pathway)
DNA polymerase (in DNA replication
example of structure function of proteins
Structure
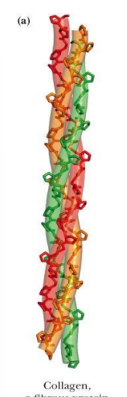
Collagen (connective tissue)
Keratin (hair, nails, feathers, horns)
example of motion function in proteins
Motion
Myosin (muscle tissue)
Actin (muscle tissue, cell motility)
example of transport function of protein
Transport
Hemoglobin (transports O2 in the blood)
Lactose permease (transports lactose across the cell membrane)
proteins are classed according to their (2)
their shape and solubility
what are the 3 categories of proteins
fibrous proteins
globular proteins
membrane proteins
what are fibrous proteins (basic structure, solubility, basic role)
Fibrous proteins have relatively simple, regular, linear structure
structural roles in cells
often water insoluble.
‘
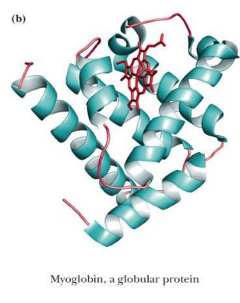
globular proteins (shape)
Globular proteins are roughly spherical in shape
ex: myoglobin
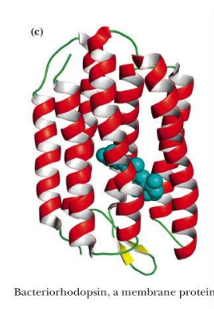
membrane proteins (found in, structure)
found in association with the various membranes of cells
fold so that hydrophobic amino acid side chains are exposed in their membrane-associated regions
primary structure is
the amino acid sequence
the sequence and composition reflect what of the protein
function
membrane proteins primarily have what type of residues
hydrophobic residues (and fewer ionic amino acids as hydrophobic does not like ionic)
fibrous proteins may have what type of sequences
atypical
Are more thermodynamically stable and have more sequences that fold into them
homologous proteins in different organisms have
homologous sequences
e.g., cytochrome c is highly conserved
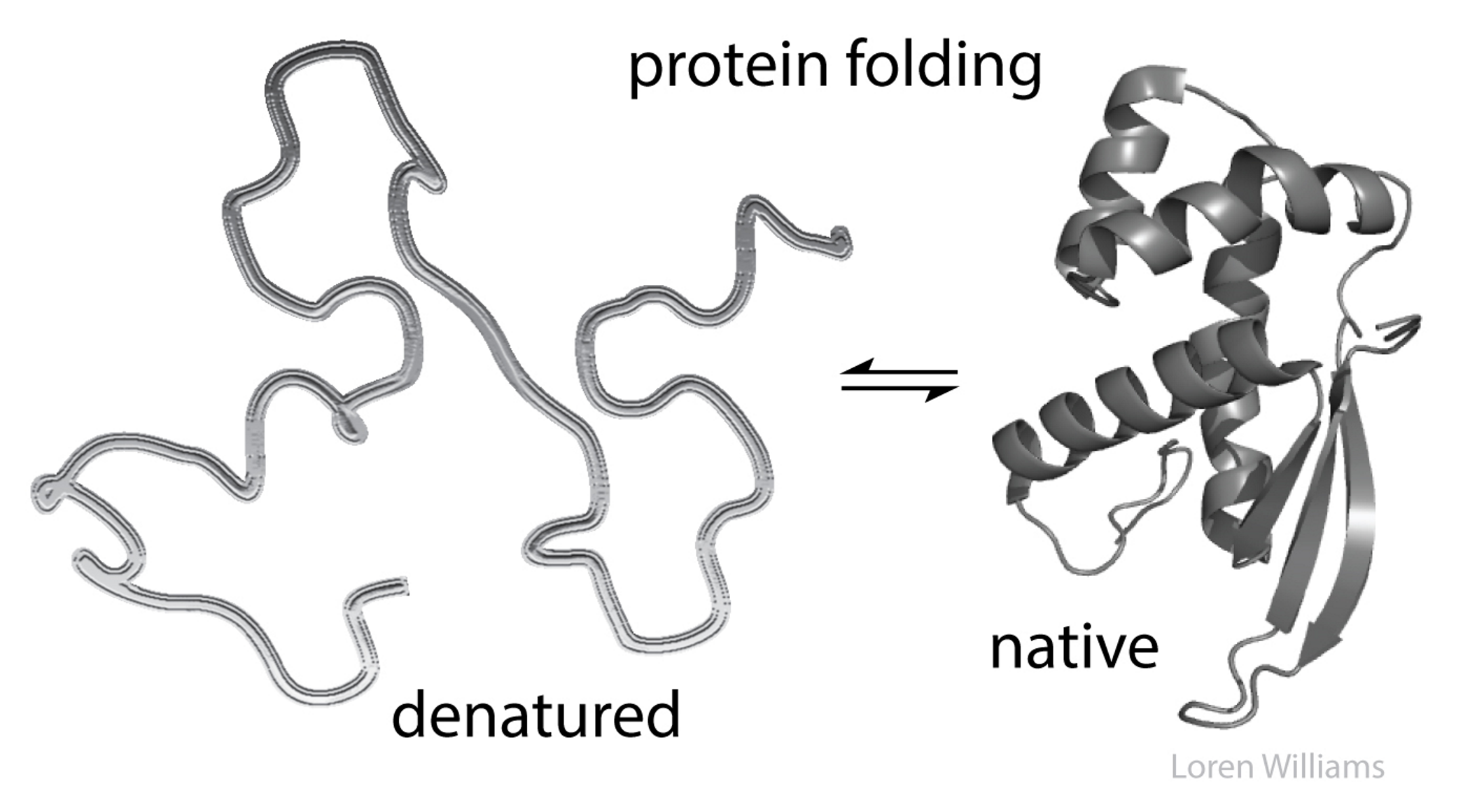
Unlike most organic polymers, protein molecules adopt a
specific three-dimensional conformation.
3D conformation of proteins entails what kinds of movement, fulfills what, is called what)
Usually entails rotation around a single bond (without breaking)
This structure is able to fulfill a specific biological function
This structure is called the native fold
what is a native fold
The functional, folded conformation (tertiary structure)
Has a large number of favorable (weak) interactions within the protein.
The H bonds, disulfide, salt bridges, hydrophobic interactions between R groups (tertiary structure)
when is protein most stable
The maximum number of weak interactions
what are the favorable interactions in proteins (weak)
hydrophobic
hydrogen bonds
London dispersion
electrostatic interactions
hydrophobic effect in proteins
Release of water molecules from the structured solvation layer around the molecule as protein folds.
The forming of hydrophobic “bonds” minimizes the interaction of nonpolar residues with water and is therefore highly favorable
hydrogen bonds (in what structures, occur where and how often)
Interaction of N-H and C=O of the peptide bond leads to local regular structures such as alpha-helices and beta-sheets
Hydrogen bonds are generally made wherever possible within a given protein structure
The backbone
Between R groups
london dispersion (is what, does what)
Medium-range weak attraction between all atoms contributes significantly to the stability in the interior of the protein
The attractive forces are due primarily to instantaneous dipole-induced dipole interactions that arise because of fluctuations in the electron charge distributions of adjacent nonbonded atoms.
a type of Van der Waal interaction
electrostatic interactions (between what groups, example of a type, seen how, where)
Long-range strong interactions between permanently charged groups
Salt-bridges, especially buried in the hydrophobic environment strongly stabilize the protein
Ionic interactions arise either as electrostatic attractions between opposite charges or repulsions between like charges
Acidic and basic R groups
N terminal and C terminal
Usually on the outside of the protein where they can interact with polar water
Salt can impact them (can get in way of attraction)
primary structure (basic def)
amino acid sequence that makes up the protein
all the information necessary for a protein molecule to achieve its intricate architecture is contained within its 1° structure

secondary structure
Secondary (2˚) structure → local areas of repeating main chain structure
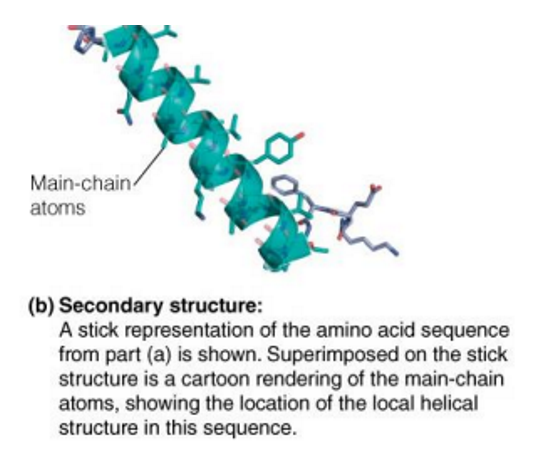
tertiary structure
Tertiary (3˚) structure → spatial arrangement of the secondary structural elements in the polypeptide chain
When the polypeptide chains of protein molecules bend and fold in order to assume a more compact three-dimensional shape
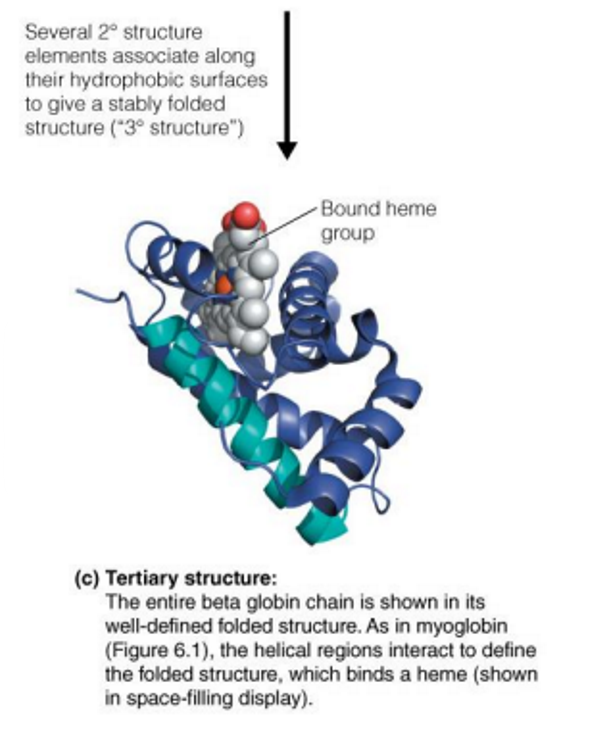
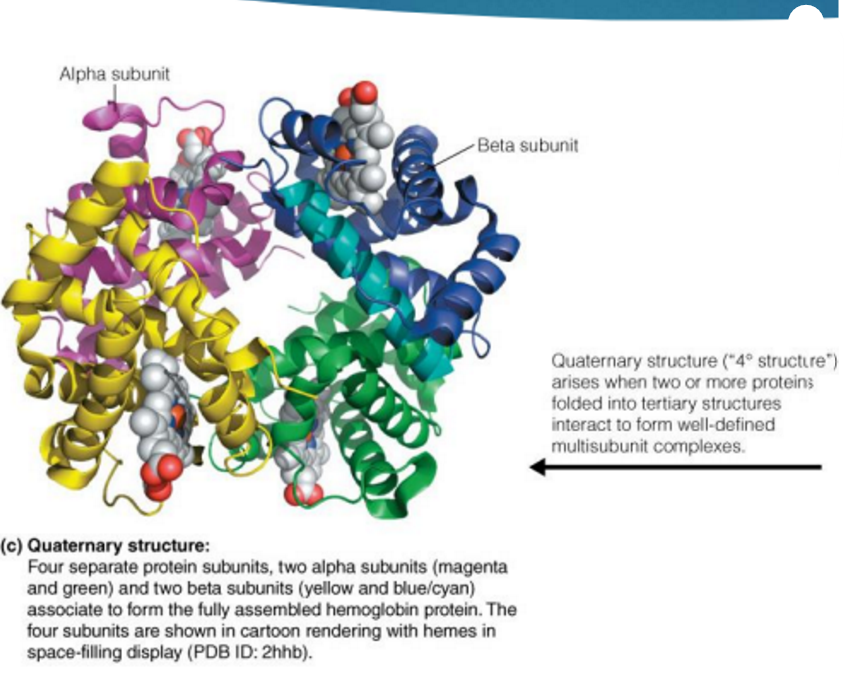
quaternary structure
Quaternary (4˚) structure → spatial arrangement of multiple polypeptide chains to form multisubunit complexes
Many proteins consist of two or more interacting polypeptide chains of characteristic tertiary structure
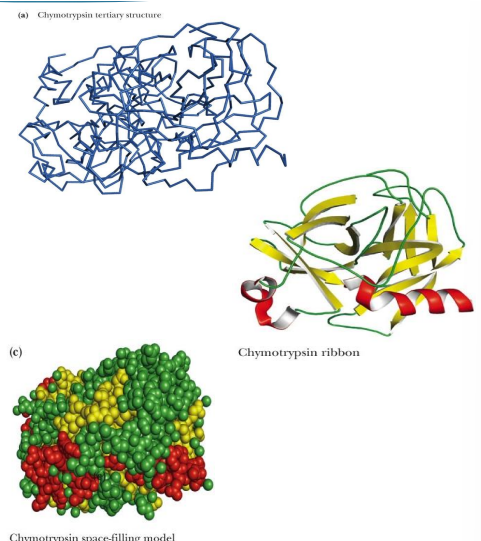
what are 4 different models of proteins (like ways they are drawn)
Backbone only
Backbone plus side chains
Ribbon structure
Space-filling structure
Each of these is an abstraction (a depiction)
conformation
A protein, or any molecule, can change its conformation by changing shape without breaking a bond
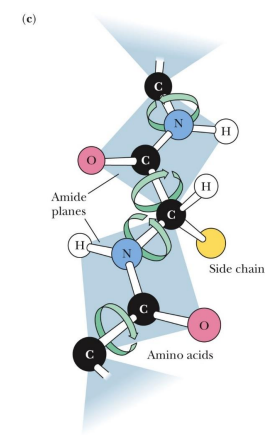
configuration
A configuration change requires the breaking of a bond.
conformation vs configuration
A configuration change requires the breaking of a bond.
A protein, or any molecule, can change its conformation by changing shape without breaking a bond
Conjugated proteins
The general term for proteins containing nonprotein constituents
If the non-amino acid part of the protein is important to its function, it is called a
prosthetic group
simple protein
Many proteins consist of only amino acids and contain no other chemical groups
glycoprotein (what group besides protein- prosthetic group)
carb group
lipoproteins (what group besides protein- prosthetic group)
lipid group
nucleoprotein (what group besides protein- prosthetic group)
RNA or DNA
phosphoprotein (what group besides protein- prosthetic group)
phosphate group
metalloproteins (what group besides protein- prosthetic group)
have metal
hemoproteins (what group besides protein- prosthetic group)
have heme group
flavoproteins (what group besides protein- prosthetic group)
have FAD or FMN
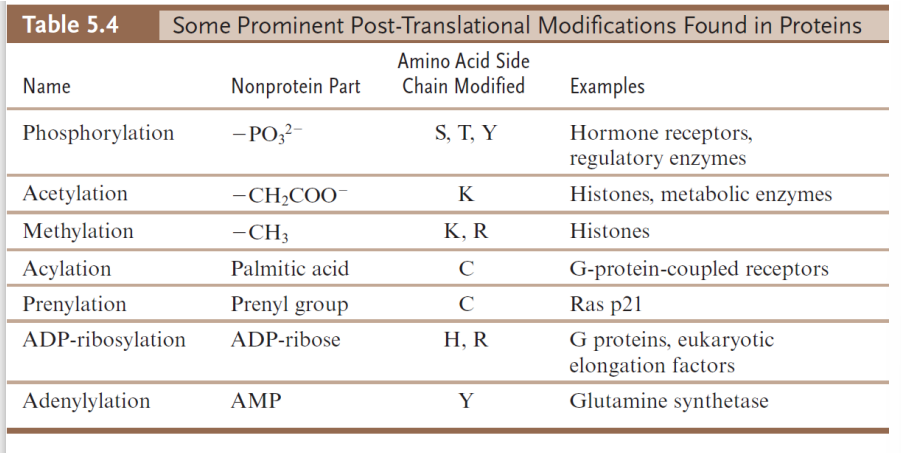
post translational modifications
chemical changes made to the protein after synthesis
Because association of the protein with the conjugated group does not occur until the protein has been synthesized, these associations are post-translational
how can proteins in a cell be isolated
basis of size and electrical charge
proteins tend to be least soluble at their
isoelectric point
At this pH, electrostatic repulsion between protein molecules is minimal and they are more likely to coalesce and precipitate out of solution
how can one use ionic strength to impact solubility of proteins
Increasing ionic strength at first increases the solubility of proteins (salting-in), then decreases it (salting-out)
Ionic strength also profoundly influences protein solubility
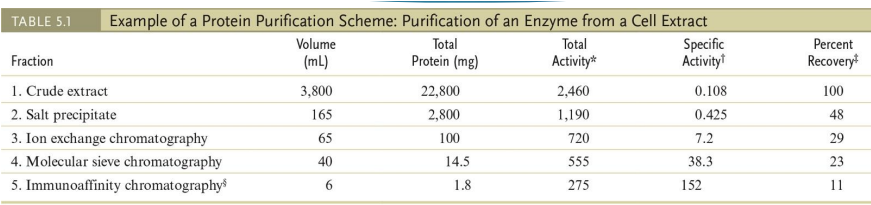
protein purification (which technique is has the highest specific activity)
A typical protein purification scheme uses a series of separation methods.
Note the dramatic increase in specific activity* of the enzyme through a series of five different purification procedures.
*The term “specific activity” refers to the activity of the enzyme per mg of protein.
electropheresis
(what is it, how it works)
Separation in analytical scale is commonly done by electrophoresis
Electric field pulls proteins according to their charge
Make all proteins negative to run towards the positive
Gel matrix hinders mobility of proteins according to their size and shape
As the protein molecules move down the gel, they experience the pH gradient and migrate to a position corresponding to their respective pI's. At its pI, a protein has no net charge and thus moves no farther
Bigger the protein the less it will travel and small will travel farther and faster

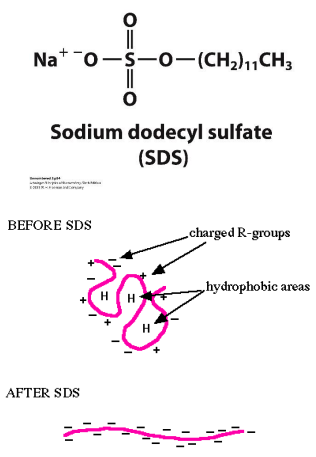
SDS PAGE: MOLECULAR WEIGHT
SDS – sodium dodecyl sulfate – a detergent
SDS micelles bind to and unfold all the proteins (tertiary structure)
SDS gives all proteins an uniformly negative charge
The native shape of proteins does not matter
Rate of movement will only depend on size:
Small proteins will move faster
SDS-PAGE is often used to determine the molecular weight of a polypeptide chain
WESTERN BLOT (what is it, what does it do, process)
AKA immunoblot
Routine technique for detecting and quantifying target proteins,
Including low levels of proteins in a complex mixture.
Can quantify but not as sensitive and accurate as an ELISA (because we are mainly looking at images not certain amount
First step is the electrophoresis
Valuable assay for identifying and understanding protein characterization elements, protein, protein interactions, modifications, and more.
the western blot method helps determine specific proteins’ presence, size, and quantity in a sample
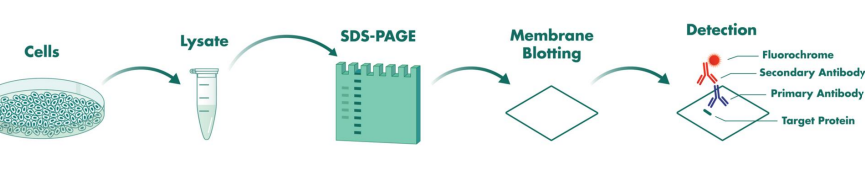
western plot process
My understanding of western blot
Get DNA and separate proteins
Run gel electrophoresis to separate proteins
Make antibody for specific protein of interest
Take gel electrophoresis and transfer onto polymer sheet to make it easier to work with
Then we add our primary antibody so that will bind to target protein
Then we made another antibody specific to the first antibody so that it binds to it (but this one allowed for color change)
Then this can now be visualized on a autoradiograph to see where exactly the antibody has bound to the protein (which band)
ELISA (what is it, what does it do, process)
Enzyme-Linked Immunosorbent Assay (ELISA)
Highly sensitive and specific enzyme immunoassay technique for quantitatively and qualitatively analyzing antibodies or antigens,
Including proteins, hormones, peptides, nucleic acids, and more.
PROCESS: An antigen from the sample is attached to a polystyrene plate during the ELISA method. An identical antibody tied to an enzyme is applied for the antigen to bind to it. After washing, all unbound antibodies are removed from the plate, and an enzyme substrate is applied after washing. When binding occurs, this enzyme substrate produces a visible signal, such as a change in colour.

types of elisa (4)
direct, indirect, sandwhich, competetive
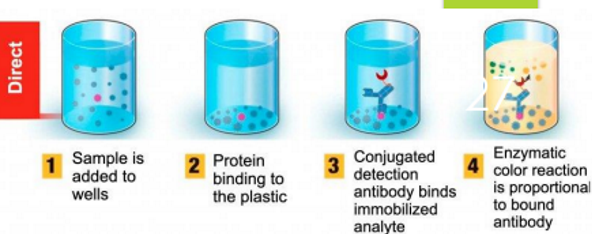
direct elisa
Direct: Here, an antigen - coated plate is used to test an antibody.
indirect elisa
Indirect : With this type, an antigen -coated plate is used to screen an antigen or antibody.
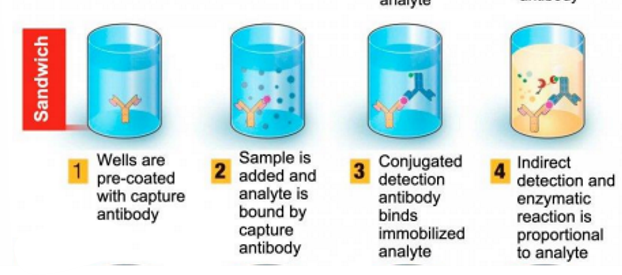
sandwhich ELISA
Sandwich : It screens antigens with an antibody -coated plate. The antigen is sandwiched between the capture and detection antibodies, hence the name.
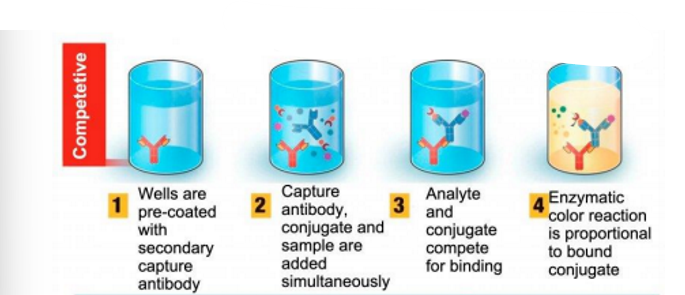
competitive ELISA
Competitive : This test is used to detect antibodies that are specific to antigens found in the serum being tested
WESTERN BLOT VS ELISA
ELISA and western blot are two popular methods for detecting and analyzing proteins
Key difference:western blot Routine technique for detecting and quantifying target proteins. ELISA Highly sensitive and specific enzyme immunoassay technique for quantitatively and qualitatively analyzing antibodies or antigens
random insert, but what is a PTM
post translation modifications (on a protein)
The full genetic potential of a cell is contained within it
genome
A more accurate reflection of what a cell is doing at any moment is found in the
proteome
because proteins are the agents of cellular function
what is proteome
where proteins are
is dynamic and may consist of hundreds of thousands of proteins,
-Result of PTMs, alternative RNA splicing, and RNA editing
proteomic tools want to
global purification strategies to separate complex mixtures,
Followed by sequence determination using mass spectroscopy to both identify and quantify each of the different proteins present
proteomic
seeks to describe the full complement of proteins present in a specific cell type under a defined set of conditions