Unit 3 - Vesicular Transport
1/55
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
56 Terms
Vesicular Transport
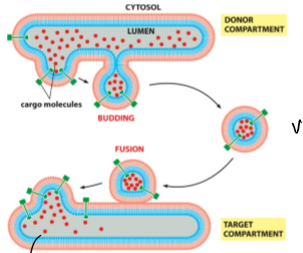
moves material between orange spaces including liquid and membrane; unstable until actually form the vesicle; 3 main coat proteins that have diff. distribution and mediate diff. transport steps, move diff. cargo, but mechanism of buddind and fusion are similiar
Vesicles
small, speherical, liquid filled, membrane-enclosed structures; move proteins/molecules between compartments; have a whole lot of proteins in their membrane (cargo, adaptor, targeting); 3 diff. coat proteins (clathrin, COPI, COPII);
FUNCTION
budding requirements - coincidence detection (right coat proteins, cargo presence) and energy (membrane deflection)
fusion requirements - coat proteins removed, spring-loaded v- and t-SNAREs, and energy required for SNARE recycling
Vesicular Transport ?’s
purpose of it, do we need distinct mechanisms to transport soluble proteins, like antibodies, and transmembrane proteins, like channels?
Coincidence Detection
cells employ multiple mechanisms to give compartments a molecular identity; only when all molecular players are present at high enough concentration do vesicles form; basically ensures vesicles form in the right way and form when/where correctly; how to achieve specificity (this and MTs); only at the correct membrane patch, do you have sufficient amounts of all of the following to form a vesicle before the timer runs out (coat proteins, phophoinositides, adaptor proteins, cargo proteins)
Vesicale Formation: COPII
Sar1 inserted into membrane but only in the cytoslic leaflet (initiates bilayer deformation); Sar1-GTP recruits inner coat (Sec23 and Sec24) and cargo proteins; coat proteins act as delayed GAPs fror Sar1-GTP; assembly begins to deform membrane and starts to look like a vesicle; more coat proteins recruited (Sec13 and Sec31); more membrane deformation; cargo proteins included; other regulatory molecules included (v-SNARE, membrane binding, membrane markers, phophoinositides); pinches off
Sar1
G-protein; bounded to GTP = membrane bound; bounded to GDP = soluble; regulated by GAPs and GEFs; recruits other proteins to make vesicle; hydrolysis of this initiates coat disassembly
Sec24 and Sec23
coat proteins that act as delayed GAPs for Sar1-GTP; way to set a timer so if cell can’t make a vesicle in that time, vesicle goes away
Sec13 and Sec31
provide more force to form vesicle; makes up outer coat; binds to Sec23/24 (binds to inner coat); can self-assemble into cages and must be removed befoer vesicle can fuse
COPII Assembly and Disassessmbly
Sar1-GTP inserts into membrane → binding of COPII inner coat proteins Sec23/24 (requires presence of cargo proteins and Sar1-GTP; also activates slow GTPase function of Sar1 → timer) → outer coat proteins Sec13/31 bind → use GTPase function as timer (at specificed time interval, Sar1-GTP becomes Sar1-GDP which causes Sec23/24 to fall off, outer coat falls off, Sar1-GDP leaves membrane) → vesicle budding then is a race between assembly and disassembly → coat must fall off of vesicles before fusion is possible
SNARE Proteins
control fusion; interaction between v (vesicle) and t (target membrane) -SNARE and from a tight bundle that pulls ends so tight they fuse; pulls vesicle close to membrane using energy (expels H2O molecules from space inbetween) from SNARE binding; unwind and recycle (NEEDS ATP - everything else is GTP driven); then repeat; like pre-wound springs that coil together and bind tightly; membranes fuse (if 1.5 nm apart, lipids can still flow from 1 membrane to another and fusing is regulated at synapse) and unwinding step recharges and recycles the SNAREs (requires protein NSF and ATP)
Recap
Vesicale Formation
Sar1 - g-protein and membrane or cytosolic; coat proteins - recruit cargo and act as delayed GAP for Sar1; coat must be removed before fusion; race between vesicle formation and time
Vesicle fusion
SNARE proteins - v-SNARE on vesicle and t-SNARE on target; act like springs to pull the membrane close
membrane fusion is biophysics
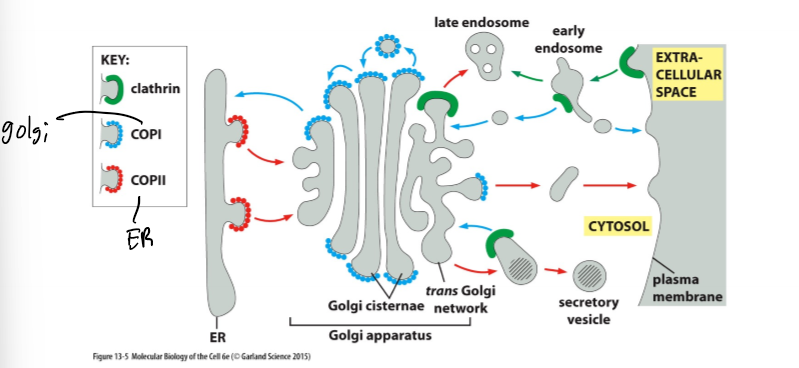
Different Coat Proteins Mediate Specific Transport Steps
Phosphoinositides (PIs)
particular phospholipid that have an inositol sugar; make up about 10% of all phospholipids in the membrane; inositol sugar is heavily modifed by phosphorylation from regulated kinases and phosphatases; the # in the PIP part tells you what position(s) it is phosphorylated at; binding proteins recognize specific isoforms; regulated localization of these isoforms (basically certian of these are in diff. spots and there are an unequal distribution of them)
Questions
How does a vesicle form? - initiation and coat proteins
How does a vesicle fuse? - v- and t-SNAREs
How does a vesicle ensure presence of cargo proteins? - coat proteins as coincidence detectors
How does a vesicle form in the right place? - co-incidence detectors, phosphoinositides
How does a vesicle fuse with the right compartment? - distribution of SNAREs, Rabs, phosphoinositides
Fusion Specificity
3 main pathways control this: distribution of SNAREs, Rab proteins, and Phosphoinositides; together they help with the origin and destination of vesicle and identity of target membrane; multiple, built-in redunancies; none of the pathways have 100% specifity but because relay on all working together, get 100% specificity
SNAREs
fusion specificity; v- and t-SNAREs are unequally distributed and only specific pairs can fuse; only a vesicle made in compartment a and ends up where it should be, will the v- and t- SNAREs fuse
Rab Proteins
fusion specificity; family of small G-proteins; Rab5-GEF activates Rab5 to become Rab5-GTP which then binds membrane; activation of PI3 kinase (which controls phosphoinositides); clustering of Rab-Effectors including tethering proteins; disassemble by using Rab5-GAP to get Rab5-GDP; recruit target proteins and will form distinct Rab domains in the membrane; present on target and on vesicle but specifc proteins may be on different on target and vesicle
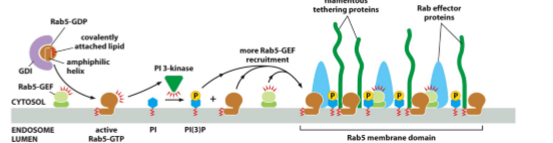
Rab Proteins continued
cycle between membrane and cytosol; held inactive by Rab-GDP dissociation inhibitor (GDI) in cytoplasm; activated by special membrane-bound GEFs; activation causes Rab to bind to membrane; membrane-bound activated effectors (tethering proteins - contribute to fusion specificity)
Phosphoinositides
fusion specificity; clustering of Rab effectors requires correct PI
Types of Vesicular Transport
regulated, constitutive, anterograde, retrograde, endocytosis, and exocytosis; move proteins and liquid throughout the cell and EC space
Regulated Transport
fast but needs receptors
Constitutive Transport
slow
Anterograde
ER → EC space
Retrograde
endosome → ER
Endocytosis
uptake of EC material; bringing in outside material; a default pathway; all material moved to endosomes/lysosomes and if not then needs an extra signal; EC space → Endosome → Lysosome
Exocytosis
sceretion into EC space; also anterograde; a default pathway; any other destination that is not the EC/plasmamembrane needs an extra signal; ER → Golgi → EC space
Organelle Functions
ER, Golgi, Endosome, Lysosome
ER
function is to do protein folding (chaperones assist) and processing; also starts glycoslayion (attachment of sugars); attaches lipids to proteins; forms disulfide bonds by protein disulfide isomerase
Glycoslylation
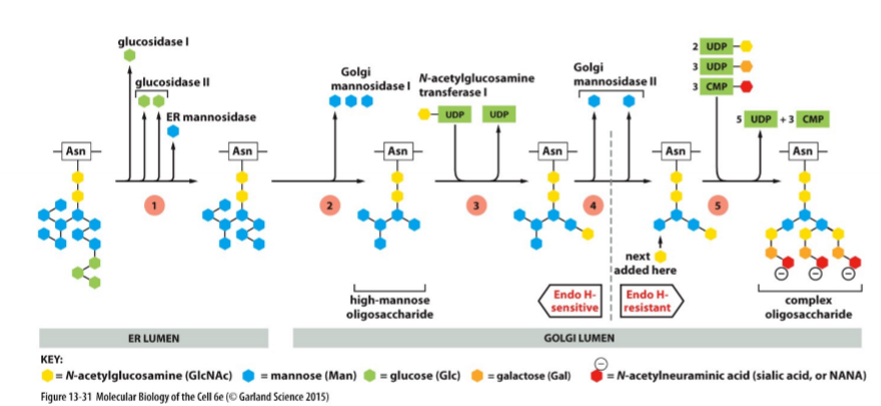
ER function; sugars transferred as a group; 2 forms - N-linked and O-linked sugars; means to hold protein inside the ER; sugars have important roles in helping proteins fold
N-linked Sugars
form of glycosylation; bind to N of asparagine - in ER; required for many proteins to fold properly but can occur on any N
O-linked Sugars
form of glycosylation; bind to O of serine or threonine - in golgi
Protein Maturation in the ER
all proteins glycoslated in ER lumen by transfer of an oligosaccharide block to N of asparagine; chaperones hold onto sugars and assist in folding; this is not always a successful process so the incorrectly folded ones are exported out of the lumen and destroyed by proteasome in the cytosol; disulfide bridges form; ER stress
ER Stress
cellular symptom; excessive levels of un or misfolded proteins; correlated with worse prognosis for certain diseases; cellular response mechanisms to improve folding efficiency and removal of misfolded items
Golgi
function is to finish gylcosylation, protein maturation, and sorting (tells things where to go); has a cis (next to ER) and trans (closest to plasmamembrane) face
Protein Maturation in the Golgi
golgi function; glycosylation - N-linked completed and O-linked started and finished; sulfation of tyrosines and sugars (adding a sulfate (- charge) so attracting water and creates a gel); modifying enzymes are markers for portions of stack; protein sorting for next step (plasmamembrane/EC space, endosome/lysosome, and peroxiosomes, etc.); all golgi proteins are transmembrane proteins
Glycosylation in Golgi
golgi function; Endo H - bacterial protein that removes the entire tree of N-linked oligosaccharides, blocked by oligosaccharide modifications in late golgi, after processing in medial golgi, glycoproteins become resistant to hydrolysis by Endo H
ER and Golgi Transport
smooth ER to cis golgi; anterograde (default secretion pathway (flow)) and retrograde transport (signal-mediated return of ER proteins that moved to the Golgi by the default transport pathway - so basically getting things back that should not have left in the first place)
Endosome
function is to retrieve material from the EC space; also sorts; cholesterol picked up by endocytosis for this
Lysosome
function is to digest biological molecules
Biological Functions of Glycosylation
chemcial
rigid, protrude away from protein, aid protein folding: rigid structure limits peptide movement, highly hydrated, can provide protective “coat”, can add - charge if sulfonated
biological
resistance of EC matrix to compressive forces, membrane protection from hydrolysis, viral membrane binding (hemagglutinin), and blood type (different sugars depending on the type)
Transport Golgi→ Plasmamembrane
constitutive and regulated exocytosis; vesicle contents released into EC space; vesicle membrane fuses to plasmamembrane; membrane fusion is often regulated (SNAREs interact and tie synaptic vesicle to membrane, yet don’t fuse and fusion occurs after recognizing AP)
Endocytosis continued
3 major forms - receptor-mediated specifc uptake, continous general uptake extracellular and transmembrane material (pinocytosis), and phagocytosis; there is an extensive exchange between extracellular and organellular fluids and between the plasma and organellar membranes; has to remain in equilibrium with exocytosis
Receptor-Mediated Uptake
form of endocytosis; specific nutrients (like cholesterol - chylomicron which is a way for your body to get this into your bloodstream) and availability of signal transduction receptors
Pinocytosis
form of endocytosis; cell drinking; happens in all cells, all the time with nutrients and lipids
Phagocytosis
form of endocytosis; takes in invading bacteria, dead/dying cells, and function of of specialized ‘housekeeping’ cells
Endosomes and Lysosomes combined Functions
endosomes (pH <7) and lysosome (pH 5) exist in a continum; lysosomes degrade cellular material; default destination for endocytosed material is the endosome and a signal is required to divert material from this path; continously fusing w/ eachother and things change all the time
Which organelle is waste removel their primary function?
lysosomes - safe disposal of biological macromolecules; have an acidic pH (ATP synthase in reverse and hyrdrolytic enzymes require this pH to function); inner membrane and proteins inside of here are glycosylated for stability; signal for lysosomal targeting of hydrolytic enzymes is a modified sugar, not a specific peptide sequence
Summary
Identification of Sec61
by Randy Schekman; knew genes that were involved in secretory pathways can be detected using genetic screens and about the N-terminal hydrophobic SS; yeast can be grown on media lacking specific aa (histidine)
Histidine
non-essential amino acid; supplied by media or synthesized in yeast by HIS4C; HIS4C and substrate usually in cytosol
Translocator Protein 1 Experiment
normal yeast can live w/o histidine in media bc can synthesize it
HIS4C removed by mutation (Mutant 1) → yeast can not survive w/o histidine in the media (it needs it in one form or another)
Mutant 2 - N-terminal SS added to HIS4C → will be in the secretory pathway
Mutant 2 can’t survive w/o histidine in the media bc you are separating the HIS4C (ER) from the substrate (cytosol)
Mutant 2 w/ mutant that abolishes translocation into ER (Mutant 3) → will survive w/o histidine bc HIS4C will be in the cytosol and the yeast will be able to synthesize it
Schekman’s Screen
was to identify mutants in peptides supposed to be going into the ER that remained in the cytosol; system was set up so interesting mutants become viable but others died; results - isolated Sec61 mutation, identified mutant gene by adding cDNAs back into yeast one at a time to identify which one reverts the phenotype, and no info on subcellular localization or molecular function of Sec61
isolating mutant ‘straightforward”, figuring out molecular nature of which gene is mutated and what its function is is difficult, and elegantly identified in vesicular transport
Does Intra-Golgi Transport exist?
yes it was discovered by James Rothman; knew they moved through the golgi but didn’t know how
Intra-Golgi Transport
mutant (donor) - no GIcNAc transferase and has a labeled viral protein
wild type (acceptor) - has GINAc transferase present but no labled viral protein;
some viral proteins that have sugars on them that Endo H could not remove so had to go from mutant to Wt golgi so yes this exists
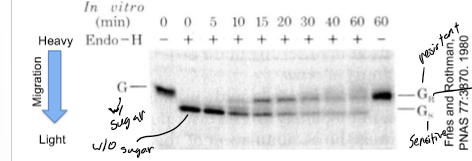
Rothman’s Experiment
infect mutant cells w/ a virus (unique protein not in WT)
mix extracts from mutant and WT cells together
is the viral protein processed by enzymes only present in WT extract (yes - material moved mutant→WT golgi; if no - material can not have moved into WT golgi)
used maturation of sugars on secreted proteins to monitor progression through golgi stack; mutant golgi - no sugar maturation (sugars removed by Endo H); WT golgi - sugar maturation (sugars not removed by Endo H);
Endo H sensitive is lighter than resistant
carrying out experiment is difficult but immediate answer; once show system works, can do elegant follow-up experiment
Scheckman vs Rothman’s Experiment
Scheckman’s isolation straightforward but molecular nature of which gene is mutated was hard and Rothman’s was difficult but w/ immediate answer; Scheckman identified molecular players in vesicular transport while Rothman’s left you with a way to do follow up experiments (isolation of NSF, SNAP, or SNAREs)