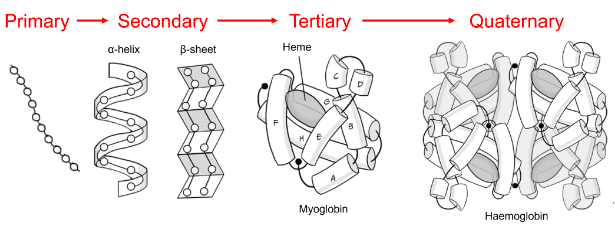
Tertiary and quaternary structure
1/43
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
44 Terms
Which of the following pairs of amino acid R groups would be capable of forming a salt bridge in a protein in a pH 7.0 buffered solution?
Glu and Arg
You need to identify the side chains that have an opposite charge at pH 7, which allows the formation of a salt bridge. Use the pKa values provided earlier.
Asp has a negatively charged R-group and His’s R-group is neutral at pH 7
Asp and Glu both have negatively charged R-groups at pH 7
Arg has a positively charged R-group and His’s R-group is neutral at pH 7
Glu has a negatively charged R-group at pH 7 and Arg has a positively charged R-group at pH 7
X is an intracellular, non-membrane bound protein. Protein Y is located within the lipid bilayer of a cell’s plasma membrane. Based on this information, how would the tertiary structure of these two proteins look like?
X has a nonpolar core with a polar outer layer, Y has a polar core with a nonpolar outer layer
Charged and polar, hydrophilic R-groups generally map to surfaces on intracellular, non-membrane bound, soluble proteins. Here, they can interact with the water. Apolar, hydrophobic R-groups tend to be buried in the cores of soluble proteins due to the hydrophobic effect.
The membrane is a phospholipid bilayer with a hydrophobic interior (due to the non-polar tails of the phospholipids) and hydrophilic exterior (due to the polar heads of the phospholipids). A protein located in the membrane generally has hydrophobic residues exposed on the surface. These hydrophobic residues are in contact with the inner, hydrophobic part of the phospholipid bilayer.
The "generally map to surfaces", and “tend to be buried” are important here; other amino acids are possible, but there is a tendency for certain amino acids on the outside or on the inside.
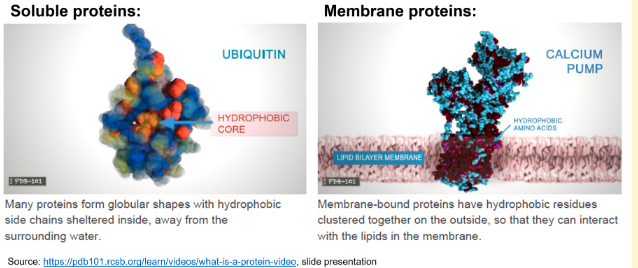
The fact that the core of most globular proteins is composed of non-polar residues is because of …
The hydrophobic effect
structural motif: a-hairpin
adjacent antiparallel a-helices
structural motif: Greek key
4 adjacent antiparallel strands
structural motif: β-α-β
2 parallel strands connected by helix
On average, the length of a super-secondary structure is around 20-50 amino acid residues, the length of a typical domain around 100-200 amino acid residues.
Proteins containing more than ~200 residues usually fold into two or more globular clusters known as domains. These domains often fold independently and have a specific function, e.g. ligand binding or enzymatic activity. So, on average, the length of a super-secondary structure is around 20-50 amino acid residues, the length of a typical domain around 100-200 amino acid residues. Exceptions exist!
The size of an average protein is ...
5 – 10 nm
Which of the following describes protein domains?
They are defined as independently foldable units
They are stable structures as compared to super-secondary structures
They are separated by linker regions
They are made up of different types of secondary structure
A homo-tetramer protein always has …
Four identical subunits
Cartoon
Space filling

Surface
Antibody
defence
Insulin receptor
communication
a-amylase
enzymatic activity
Calcium pump
transport
Ferritin
storage
Collagen
structure
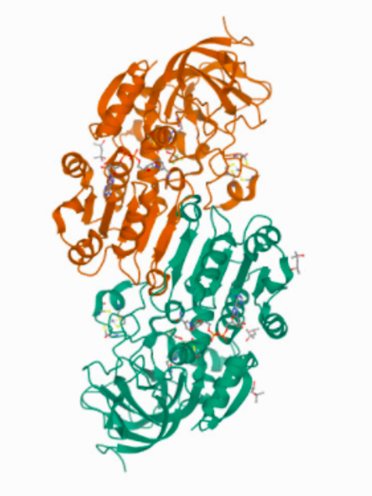
4DXH, the structure of horse liver alcohol dehydrogenase
4DXH represents the structure of horse liver alcohol dehydrogenase complexed to the cofactor NAD+ and an unreactive form of the substrate
Overall, 4DXH is a homo -2- mer, with 4 Zn2+ ions bound
The subunit-subunit interface is comprised of an antiparallel β-sheet.
Glycine and proline are the most abundant amino acids in the structure of ...
collagen
Collagen…
has a very regularly repeating primary protein structure
Substitution of glycine in the primary sequence of collagen with almost any other amino acid results in defective collagen because…
the contact points of the triple helix are so close that only glycine can fit well in the space available
The collagen defect present in scurvy is…
decreased protein stability due to decreased hydroxylation of proline residues
The quaternary structure of human haemoglobin is best described as a…
tetramer of 2 different subunits
The molecular mass of myoglobin is about __ % that of haemoglobin.
25%
On a Ramachandran plot the entries for haemoglobin would be clustered around…
the right-handed a-helix conformation
haemoglobin is a hetro-tetrameter
all of haemoglobin’s subunits are very similar to myoglobin, which is formed from 8 right-handed a-helices surrounding an iron-containing heme
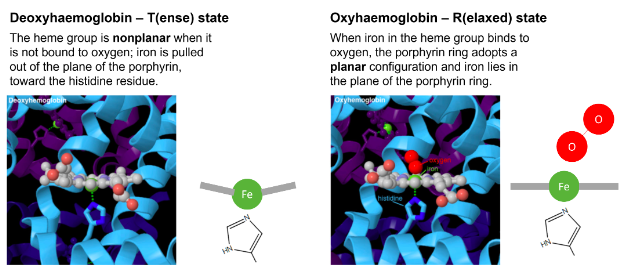
In deoxy haemoglobin, the Fe(II) is coordinated to ...
4 nitrogens of heme and to one His residue in haemoglobin
in deoxy-haemoglobinm four of the coordinated sites of iron are occupied by nitrogens of the porphyrin ring
the fifth site is occupied by the proximal histidine
the heme group is non-planar when it is not bound to oxygen
deoxy-haemoglobin is said to be in T-state (tense)
in oxy-haemoglobin, the sixth coordinated position of iron is occupied by oxygen
the porphyrin ring now adopts a planar configuration, and the iron lies in the plane of the porphyrin ring
it is also referred to as R-state (relaxed)
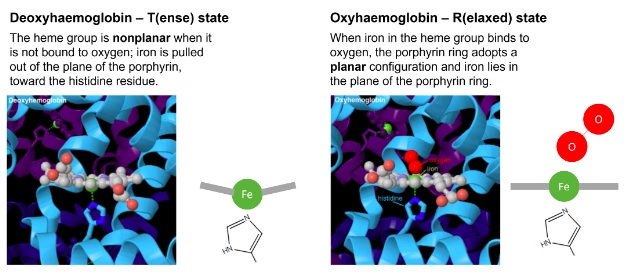
What happens when a haemoglobin subunit binds oxygen?
a histidine residue changes position within the subunit
in deoxy-haemoglobinm four of the coordinated sites of iron are occupied by nitrogens of the porphyrin ring
the fifth site is occupied by the proximal histidine
the heme group is non-planar when it is not bound to oxygen
deoxy-haemoglobin is said to be in T-state (tense)
in oxy-haemoglobin, the sixth coordinated position of iron is occupied by oxygen
the porphyrin ring now adopts a planar configuration, and the iron lies in the plane of the porphyrin ring
it is also referred to as R-state (relaxed)
What happens when a haemoglobin molecule binds to oxygen?
the second oxygen molecule binds more easily than the first oxygen molecule
oxyhaemoglobin is created which has a bright red colour
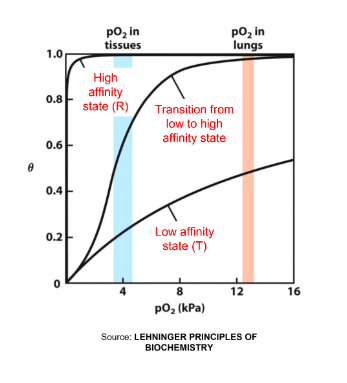
The cooperativity of oxygen binding to haemoglobin results in a…
higher affinity for the last oxygen bound than for the first
extensive protein conformational change
sigmoidal binding curve

When the phosphorylatable tyrosine of the insulin receptor is mutated to ___ the cells are unable to take up glucose from the bloodstream. When the phosphorylatable tyrosine of the insulin receptor is mutated to ____ the cells show a constant uptake of glucose from the bloodstream.
Ala
Asp
Phosphorylatable residues such as serine, threonine, or tyrosine are often replaced for phosphomimetic aspartic or glutamic acid or phosphor-deficient residues, such as alanine.
Serine, threonine or tyrosine are not charged, they do have a hydroxyl group that can be phosphorylated. When they are phosphorylated, they are negatively charged.
When we mutate them to alanine, this position in the protein cannot be phosphorylated as alanine does not contain a hydroxyl group.
When we mutate them to aspartic or glutamic acid, this position in the protein carries a negative charge, which might be perceived by the cell as a phosphorylated residue.
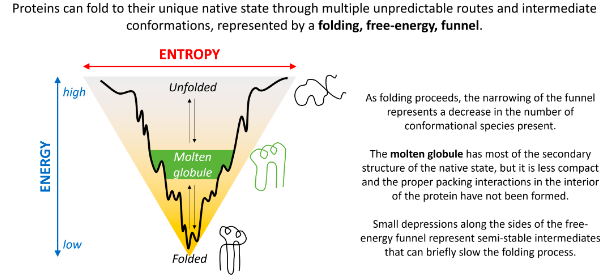
Spontaneous folding of proteins
it may be defective in some human diseases
it may involve a gradually decreasing range of conformational species
it may involve initial formation of a highly compact state
it may involve initial formation of local secondary structure
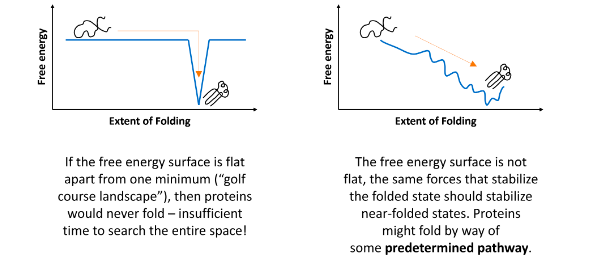
Anfinsen’s experiments on denaturation and renaturation after the reduction and oxidation of the S-S bonds in the enzyme ribonuclease provided the first evidence that the amino acid sequence of a polypeptide chain contains all the information required to fold the chain into its native, 3D structure.
Anfinsen's classic experiments on ribonuclease demonstrated that after denaturation (unfolding) and subsequent renaturation (refolding) under appropriate conditions, the enzyme regained its native, functional 3D structure. This provided clear evidence that the primary amino acid sequence of a protein contains all the necessary information to direct its folding into the correct, biologically active conformation.
What conditions are important for protein folding?
pH
temperature
Cellular milieu - the crowded environment of the cell, along with chaperones, redox conditions, and other factors, plays a crucial role in proper folding
At the midpoint of a temperature denaturation curve…
half of the proteins are denatured
Keq = 1
ΔG0 = 0
[Native] = [Unfolded]
The melting temperature (Tm) of a protein..
.. is the temperature at which the protein is 50% unfolded
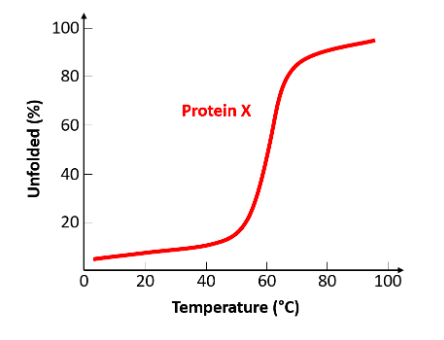
Using the sketched diagram, what is the melting point of protein X?
60°C
The interactions of ligands with proteins are…
specific - ligand binding typically involves specific interactions, such as hydrogen bonds, ionic bonds, and hydrophobic interactions, which occur at defined binding sites.
transient - ligand-protein interactions are often transient, allowing the ligand to bind and unbind as needed for biological processes (e.g., enzyme-substrate interactions or oxygen binding to hemoglobin).
Once a ligand dissociation constant (Kd) has been determined it is possible to calculate…
the ligand binding constant (Ka)
ΔGo for the binding interaction
the concentration of ligand required for half-maximal occupancy
Kd = 1/Ka
ΔGo = -RT ln(Kd)
The enthalpy (ΔH) cannot be determined from Kd
When the fraction of bound receptor – or occupied binding sites (Θ) = 0.5, [L] = Kd
Receptor-ligand binding
the larger the ligand binding constant (Ka), the smaller the ligand dissociation constant (Kd)
If the dissociation constant Kd is 10 mM, which value below best approximates the concentration of ligand at 10% saturation?
1 mM
If the dissociation constant Kd is 10 mM, 1 mM best approximates the concentration of ligand at 10 % saturation, i.e. when 10 % of receptor is bound or when θ = 0.1. It comes from the following calculations/approximations:
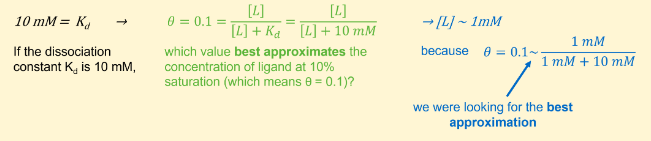
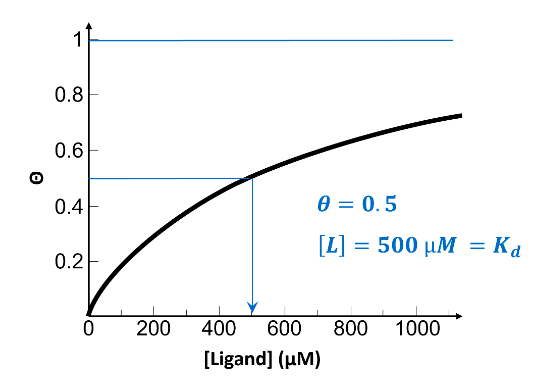
From the information presented on this graph, select the best approximate value for the dissociation constant, Kd, for this receptor-ligand system.
500 µM
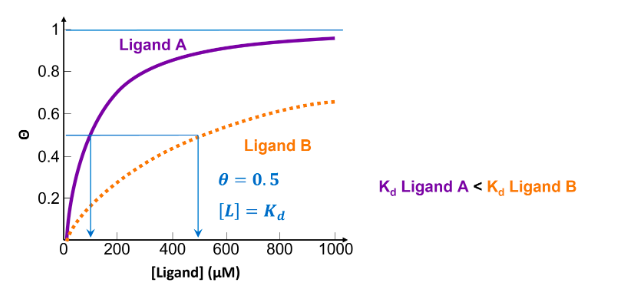
![<p>Below is a plot of θ versus [L] for two ligands. Which ligand has the lowest affinity?</p>](https://knowt-user-attachments.s3.amazonaws.com/1fbcfb75-6f4a-41f9-a0e5-7a8bab1e23da.png)
Below is a plot of θ versus [L] for two ligands. Which ligand has the lowest affinity?
Ligand B
the lower the value of Kd, the less ligand required to achieve 50% occupancy, indicating a higher affinity between receptor and ligand. Ligand B has a higher Kd, so a lower affinity.