C.Bio Endoplasmic Reticulum
1/28
Earn XP
Description and Tags
It's about the endoplasmic reticulum!
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
29 Terms
WHO ISSSSSS THE ER?
The biosynthetic factory of the cell. Only proteins that cross membranes are made in here.
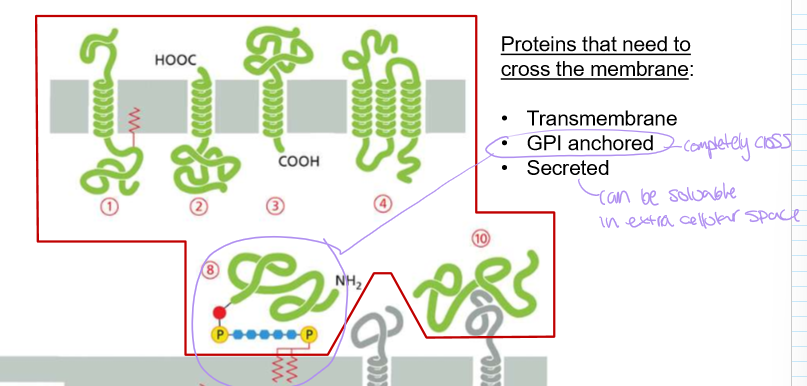
What kind of proteins need to cross the membrane?
Transmembrane (portion in membrane)
GPI Anchored (completely crosses membrane)
Secreted (often soluble in extracellular space)
Who discovered the biosynthetic pathway of protein secretion pathway and HOW (brief)
George Palade, who tracked pancreas protein secretion via pulse-chase experiments
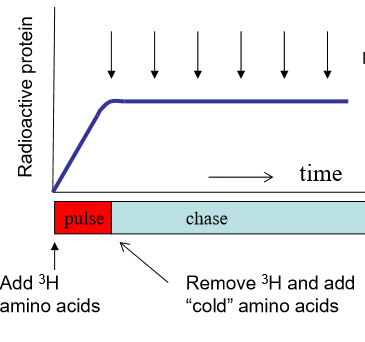
Explain the pulse chase experiment’s methology
Basically, you can label whatever you want to track. This is the pulse. Then chase it with normal objects of interest. Overtime, track the movement of your labeled thing. In terms of amino acids for protein synthesis:
first add radioactive 3H to the culture to make radioactive amino acids (pulse)
Remove 3H after a short time, and then add normal amino acids (chase)
Watch proteins migrate over time.
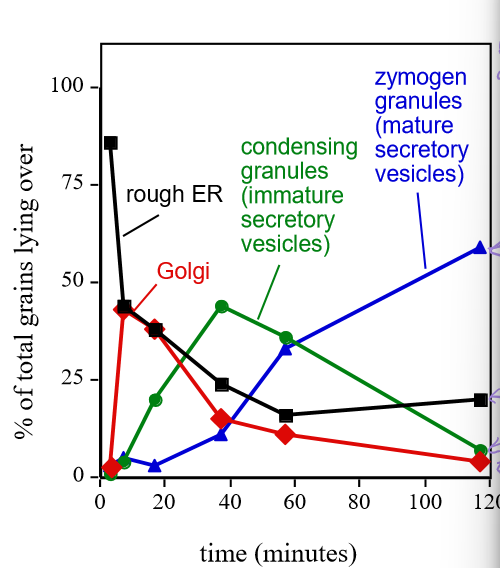
Describe Georgy Porgy’s results
Proteins initially started in the ER, but overtime, they would peak in different locations as the proteins were migrated via secretory vesicles to different parts of the cell. Final location is where the concentration continues to increase.
What are the location hotspts (7) for transmembrane & luminal protein synthesis?
Rough ER
Smooth ER (exit sites)
ERGIC (ER-Golgi intermediate compartment)
Cis-Golgi
Trans-Golgi
Secretory vesicles
Plasma Membrane/extracellular spacw
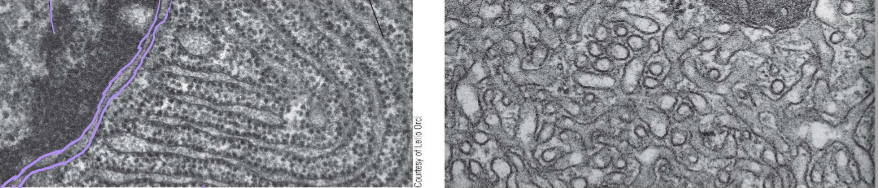
Identify these images
Left: Rough ER next to the nuclear envelope
Right: Smooth ER exit sites
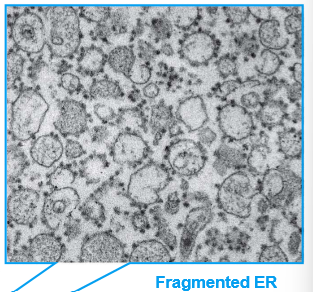
Explain the methodology of Cell Fractionation (By George Paladae AGAIN)
Cell homogenate is centrifuged many times at increasingly high speeds until microsomes and small vesicles can be obtained in the pellet.
What is in each pellet at each level of centrifugation of cell homogenate? Recall that only supernatant gets carried to the next centrifuging state
Low Speed Centrifugation: Pellet→ whole cells, nuclei, and cytoskeletons.
Medium SC: Pellet→ mitochondria, lysosomes, peroxisomes
High SC: Pellet→ microsomes, small vesicles. WE SNATCH THESE FAST
Very High Speed: Pellet→ ribosomes, viruses, large macromolecules
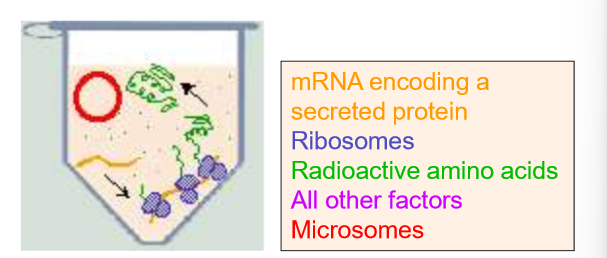
Explain what the Translation in Cell-Free system helped us to determine.
Which proteins were actually secreted in the ER for the ER lumen.
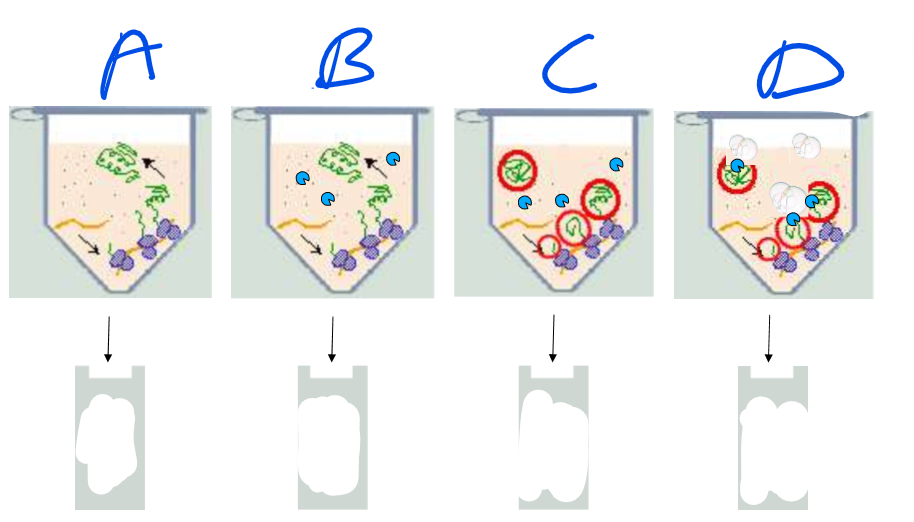
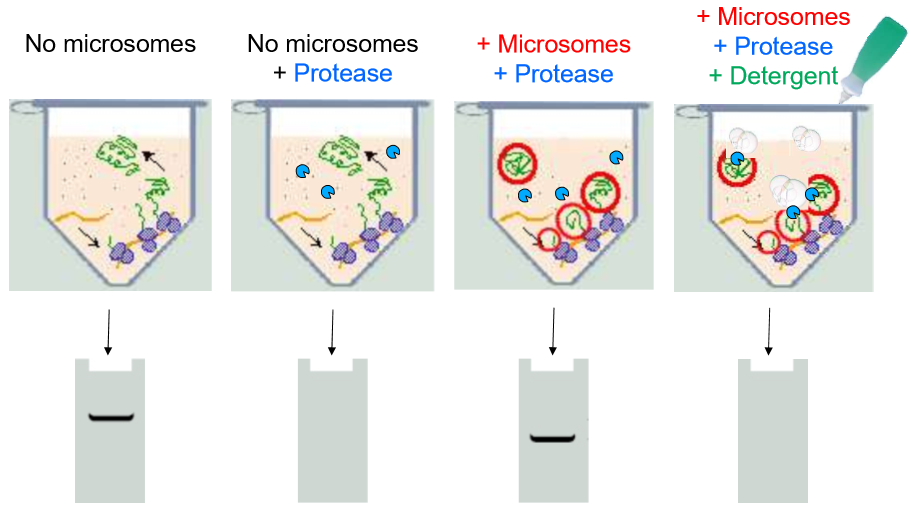
Explain each column of this experiment
Overall: this is monitoring the translation of a suspected ER protein
A: No microsomes are added
the ribosomes simply create free-floating proteins. Controls for ribosome, mRNA, and protein-folding functionality
They show up in the gel plate as longer sequences (closer to the electrode)
B: No microsomes, yes proteases
All the proteins are degraded
no proteins in the gel
C: Microsomes and Proteases
The proteins appear to be synthesized in the ER rather than in the free floating juices. Proteases cannot degrade them.
Proteins in gel are shorter and still present, showing a signal that locates the proteins is cleaved but proteins are maintained
D: Microsomes, protease, and detergent
Detergent disrupts microsomes
Proteases enter and destroy proteins
No proteins in gel, proves that proteins were inside ER lumen. Also insures that it wasn’t some inhibition of the proteases caused by the presence of the microsomes
Conclusions:
Secretory proteins are translocated into the ER lumen
in the process, they lose the N-terminus signal region of their sequence
What is the signal that indicates a protein should be imported to the ER?
A long hydrophobic stretch near the N-terminus
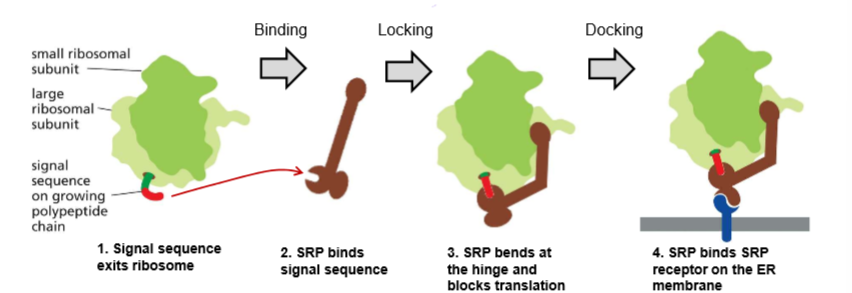
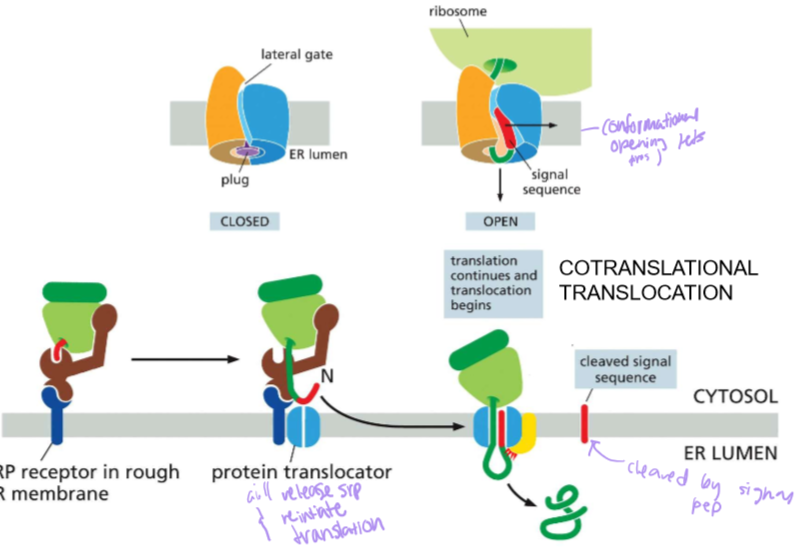
Who is SRP?!?
Signal Recognition Particle:
SRP binds to the signal sequence that exits the ribosome to pause translation before binding the ribosome and the translating protein onto the SRP receptor in the ER membrane
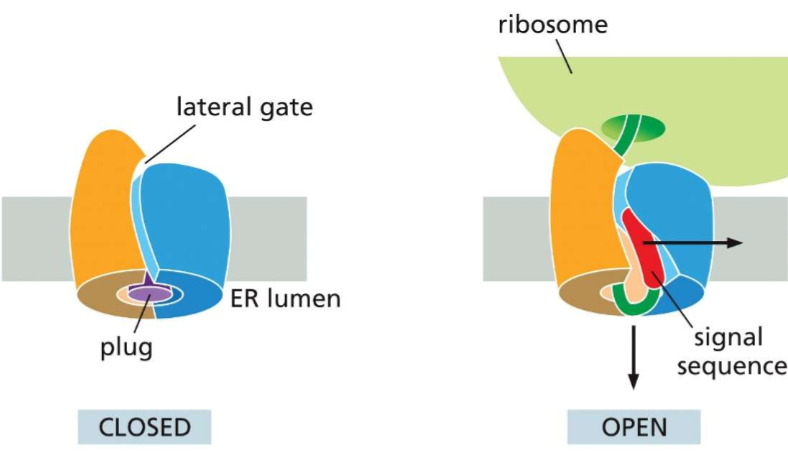
Who is Sec61?!?
This three subunit protein is THE protein translocator. After SRP docks into it’s receptor, the translating protein enters Sec61’s lateral gate and will continue translating the protein.
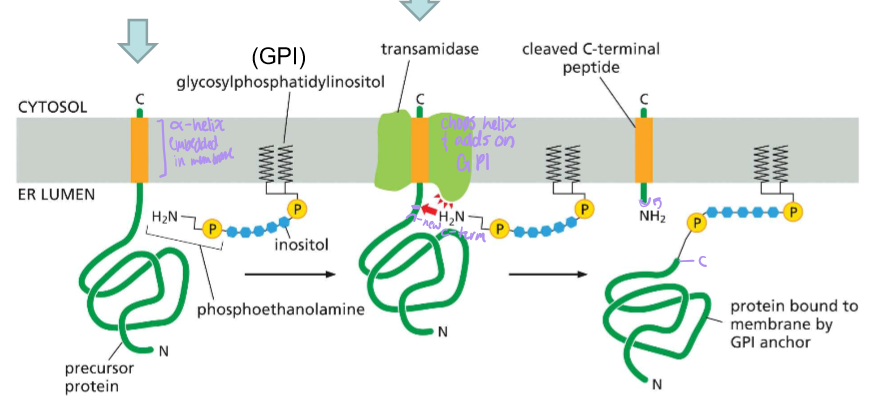
Detail the steps, post sec61 translocator, for the synthesis of GPI-anchored proteins
Post translocator protein, the precursive protein is still bound by a hydrophobic alpha helix signal embedded in the membrane
Transamidase, which is present in the membrane along with glycophosphatidylinositol, cleaves the C-terminal signal and attaches GPI.
Where is the hydrophobic signal sequence on a luminal ER protein
Near the N-terminus
Where is the hydrophobic signal sequence in a GPI anchored ER protein?
Near the C-terminus
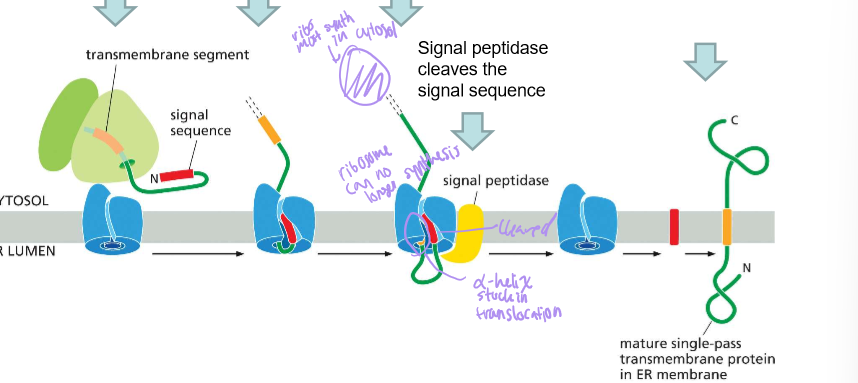
Where is the hydrophobic signal sequence of a single-pass WITH a signal sequence?
Near the N-terminus
Describe the steps of a single pass membrane protein with an amino terminus signal sequence
Signal sequence targets the protein translocator and initiates translocation
The transmembrane domain enters the translocator and stops translocation
The signal sequence is cleaved by signal peptidase
Then the protein leaves the translocator as synthesis of the protein then continues in the C-terminus of the cytoplasm.
Describe the steps of a single pass membrane protein with NO amino terminus signal sequence
The transmembrane domain is simply recognized by SRP when it leaves the ribosome.
Sec61 will orient them into the membrane of the ER.
If the whichever side of the protein has more positive amino acids, will face outwards towards the extra cellular fluid. This is because the outside membrane of the ER is negatively charged
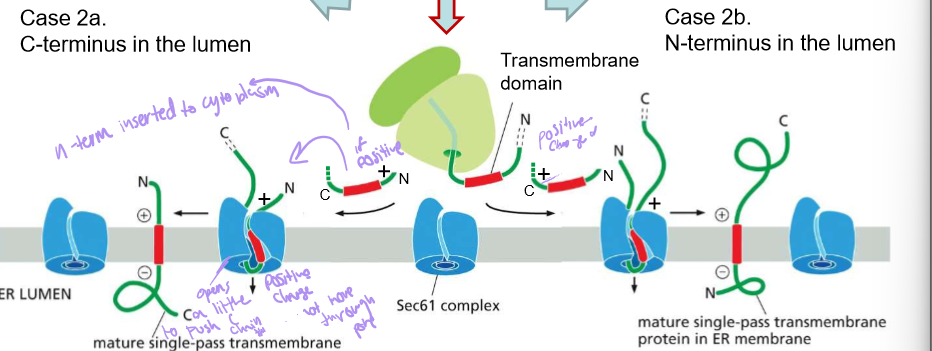
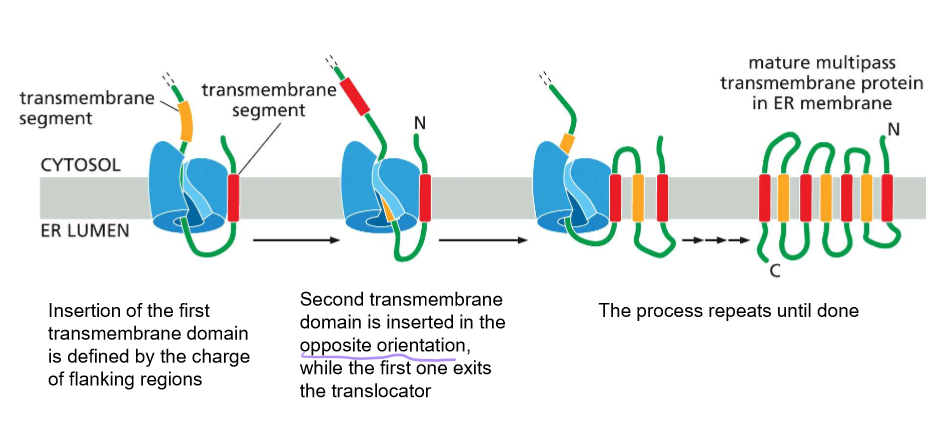
Describe the synthesis of multi-pass proteins
Note: SRP is needed for the insertion of each passing helix.
insertion of the first TMD by sec61 is determined by the charge flanking amino acids
The second TMD (transmembrane domain) is inserted in the opposite direction when the first exits the translocator.
Repeat until completed
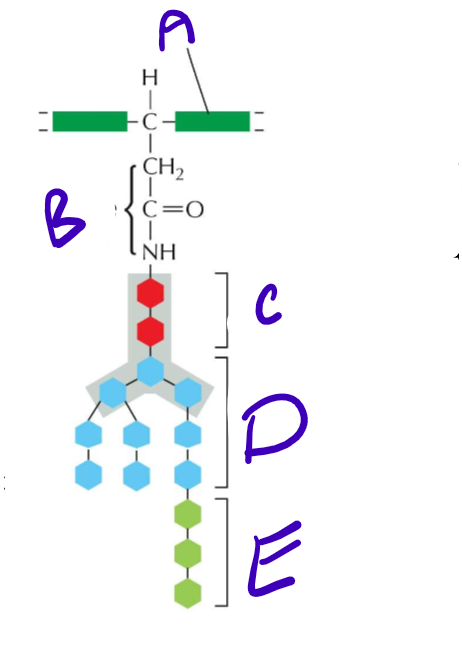
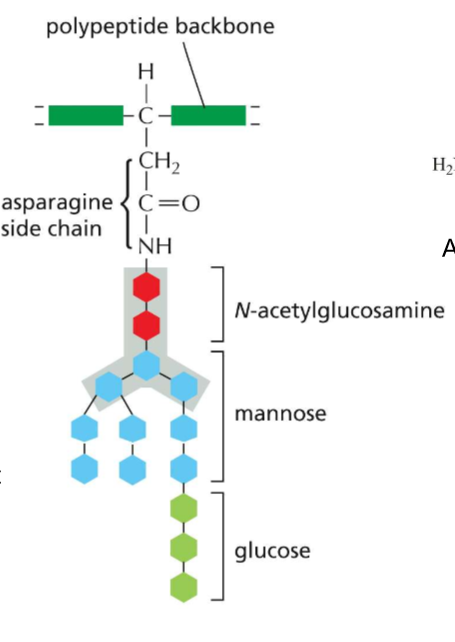
LABELLLLL Structure of an N-Linked precursor oligosaccharide!! Also what is her purpose?
A) Polypeptide backbone
B) Asparagine’s R group
C) N-Acetylglucosamine
D) Mannose
E) Glucose
She signals that the protein has properly folded in the ER
Who are the chaperones of quality control for ER protein folding?
Calnexin and some other babies
Detail the steps for quality protein folding
Unfolded protein has some glucose trimmed off its precursor oligosaccharide
Calnexin and other chaperones hold the protein to prevent aggregation as it tries to fold.
Once done, glucosidase splits the final glucose off the protein.
If 2 is not successful, rather than leaving the ER, glucosyl transferase puts glucose back on the protein and forces it to undergo the process again
What is protein aggregation?
How prions are formed… in all realness, it’s when the hydrophobic parts of seperate proteins clump together instead of folding properly, creating unusable proteins that can go on to clump and aggregate others.
What does the Smooth ER do?
Synthesize lipids.
WHERE IS FLIPPASE AND WHAT DOES SHE DO?!?!
In the golgi and other cell membranes, it ensures that specific phospholipids stay on each monolayer. This can result in the asymetric growth of the PHOSPHOLIPID BYLAYER!!
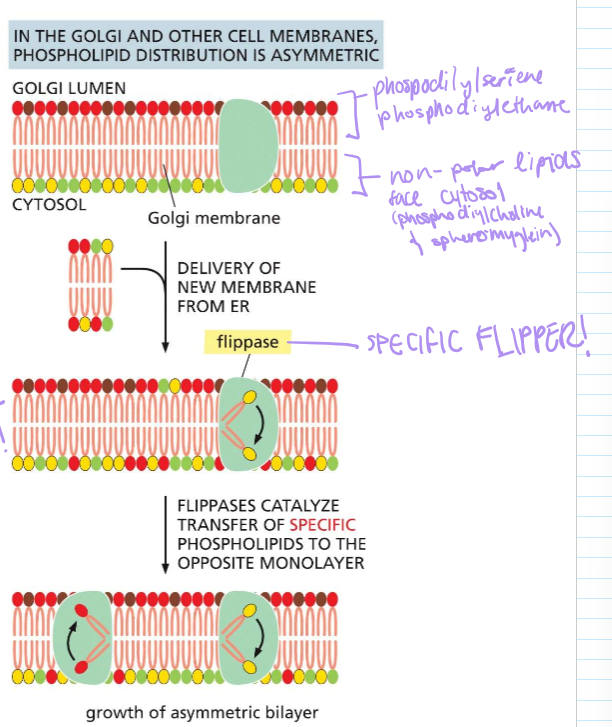
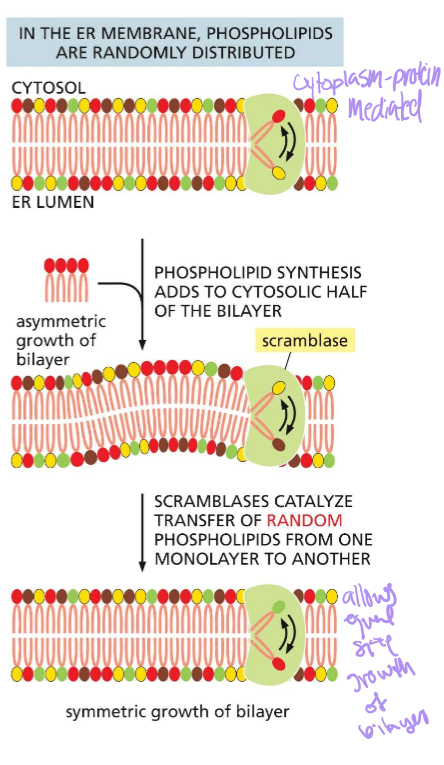
WHERE IS SCRAMBLASE AND WHAT DOES SHE DO?!?
Mediates random phospholipid distribution in the ER membrane. When phospholipids are asymmetrically added, scramblase will flip them until the numbers are even.
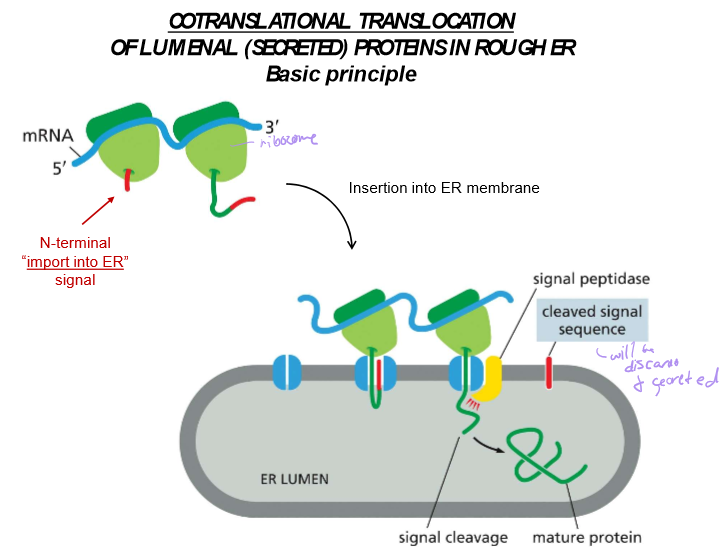
Describe cotranslational translocation
The translation of a protein into the ER due to an import signal.
In most cases, the signal is near the n N-terminus. In other instances, it may be elsewhere in the mRNA code. In all instances, proteins are translocated into the membrane by the Sec61 Translocator.