Apoptosis: Cell Cycle
1/29
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
30 Terms
ced-9 (gf) was mapped to a small region b/w markers X and Y. This region contains 15 genes. What methods can be used to identify Ced-9
sequence DNA from ced-9 (gf) to determine which gene contains a mutation
use RNAi to knock down each gene in WT worm to determine which RNAi can increase cell death
Not correct (in this case):
Isolate clones for each gene from a genomic library of wild type worms and transform them into ced-9 (gf) to determine which one can produce a wild type phenotype.
It won’t work here because the mutation in question is a gain of function with a dominant phenotype. Introducing a copy of the WT gene will not complement the activity of a gene that has more activity than normal.
Ced-9 is a homolog of the human gene Bcl-2
Homolog: genes that related b/c they have descended from the same ancestral gene
ced-9 was isolated by positional cloning
ced-9 encodes a protein similar to Bcl-2 (in humans) [29% identity at amino acid level)
BcI-2 inhibits cell death in humans – similar function as Ced-9 in C. elegans
Bcl-2 can partially complement cell death in a ced-9 (lf) mutant → conservation of cell death pathways b/w species
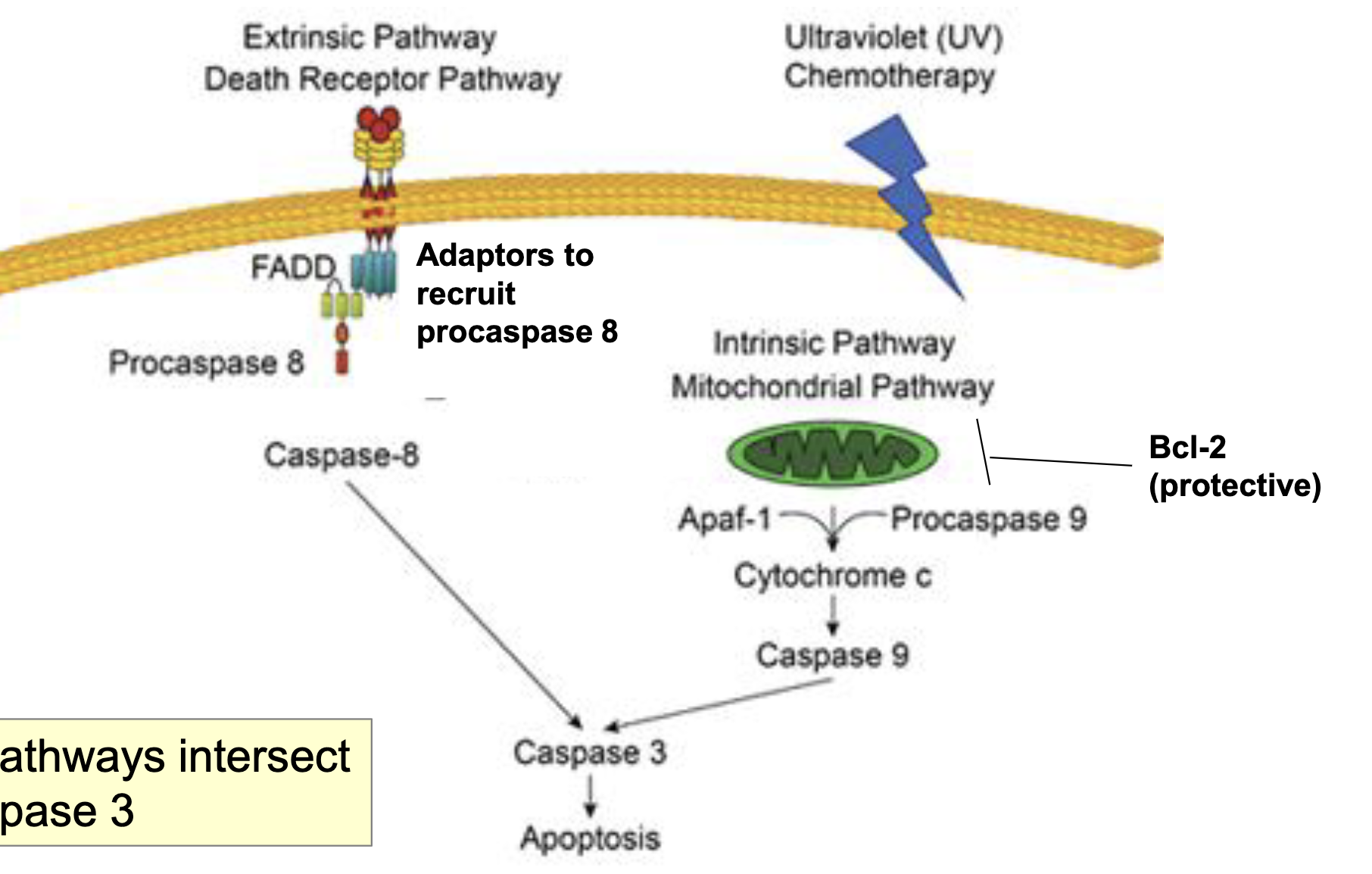
Pathways intiating Apoptosis
activation of cell death from inside the cell (intrinsic/mitochondrial pathway)
activation of cell death from outside the cell (extrinsic/death receptor pathway)
extrinsic → signaling thru receptor → apoptosis
these two pathways intersect at the effector caspases (causes events of cell death)
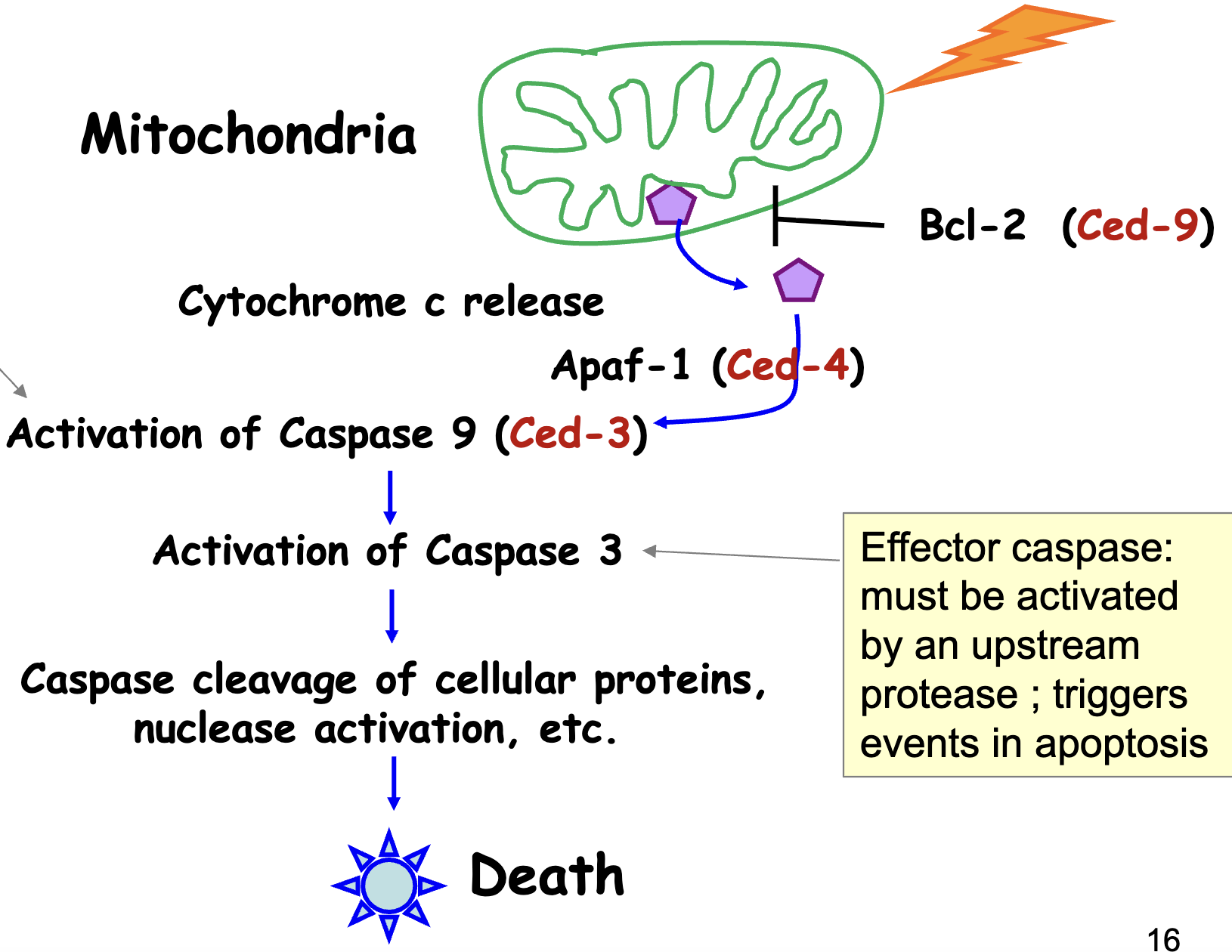
Intrinsic/Mitochondrial Pathway
when there’s an event that triggers apoptosis → cytochrome C release → forms complex w/ Apaf 1 + caspase 9
initiator caspase 9: self-activates when compexed w/ cytC and Apaf-1
once caspase 9 is activated, it processes caspase 3
effector caspase 3: must be activated by an upstream protease; triggers events in apoptosis
can’t be self-activated
Bcl-2 protects integrity of mitochondria
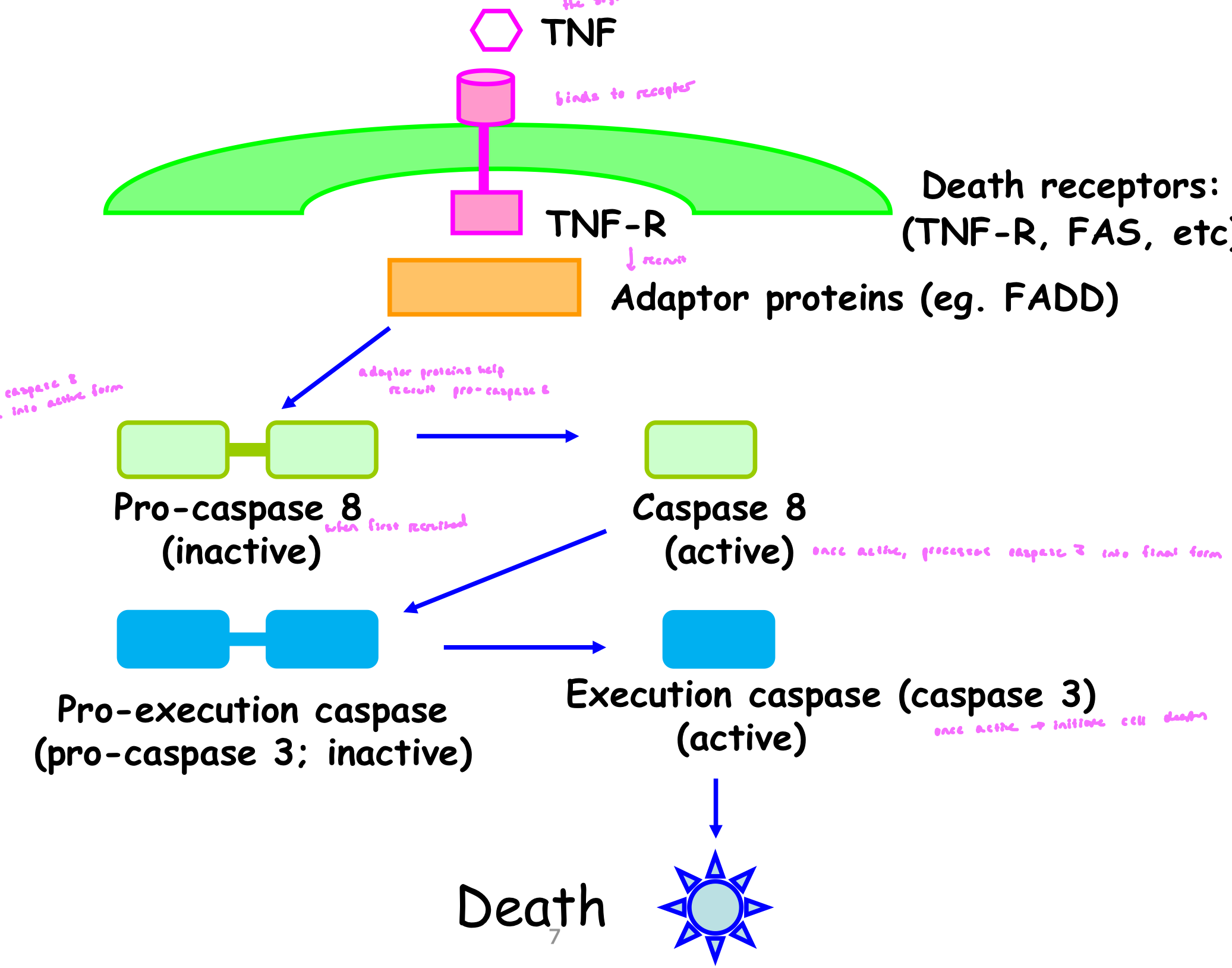
Extrinsic/Death Receptor Pathway
signal binds to death receptor
receptor recruits adaptor proteins
adaptor proteins help recruit pro-caspase 8 (inactive when first recruited); recruitment allows caspase 8 to self process into active form
once caspase 8 is active, it processes caspase 3 into final form
Similarities b/w pathways
both intersect at caspase 3
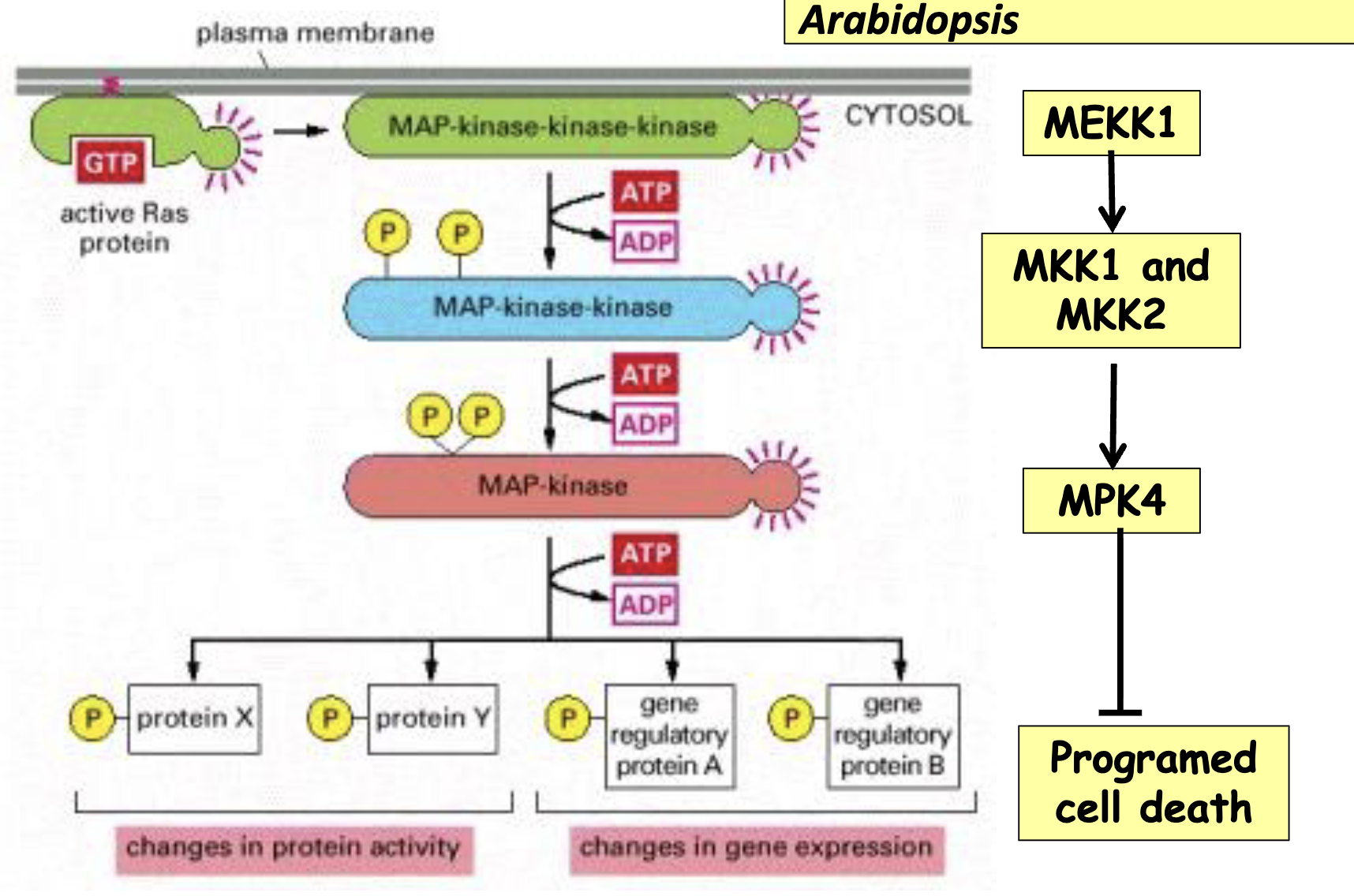
Cell Death in Arabidopsis: Background
MEKK1, MKK1/MKK2 and MPK4 form a MAP kinase cascade to inhibit programmed cell death in Arabidopsis
in a mekk1 lof, we’d expect to see increased cell death (can’t activate MKK1/2 → can’t activate MPK4 → more death)
all 3 of these promote cell survival
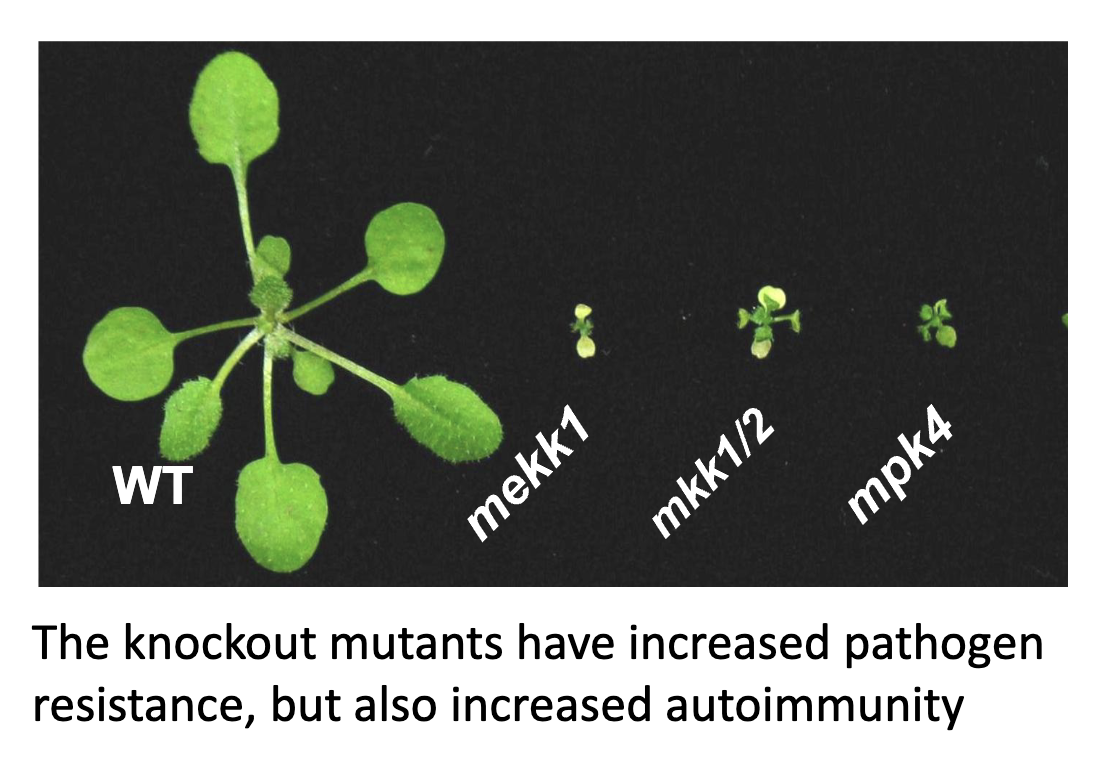
Single Mutants of MEKK1, MKK1/2, MPK4
MKK1 and MKK2 have overlapping functions, if you knock out one, the other can still compensate
would need to knock out both to see the single mutant phenotype
single mutants have a lot more cell death
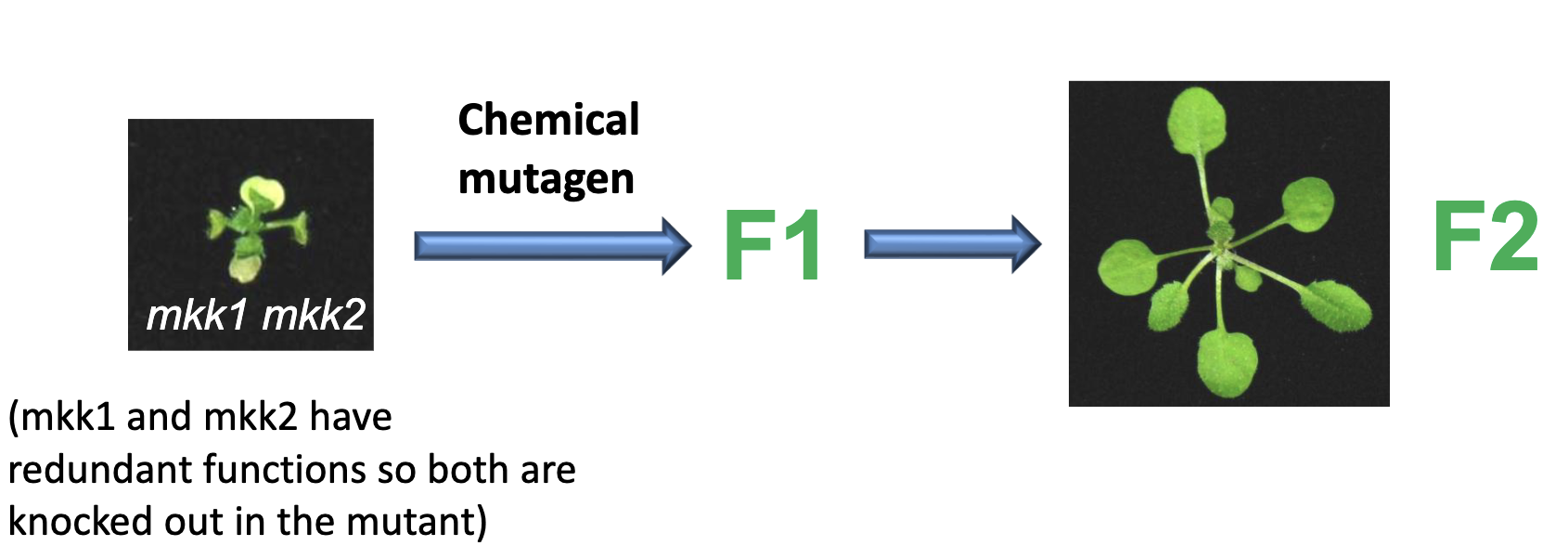
Suppressor Screen: mkk1 mkk2
finding mutation that reverse this mutant phenotype must act in the same pathway
means we got another mutation in another gene in the same pathway
mutations identified: summ = suppressor of mkk1 mkk2
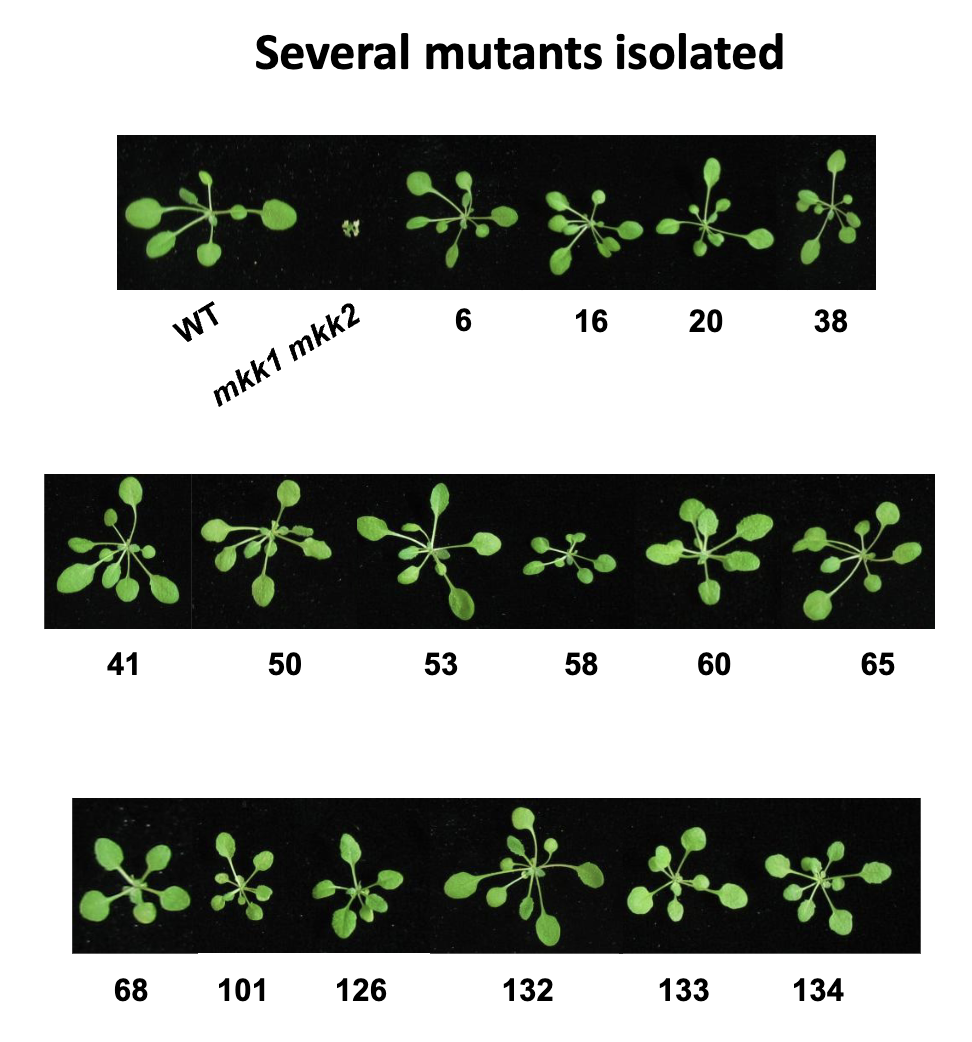
Isolated Suppressors of mkk1 mkk2 (summ)
several mutants isolated
cell death in mkk1 mkk2 is suppressed by summ2-1
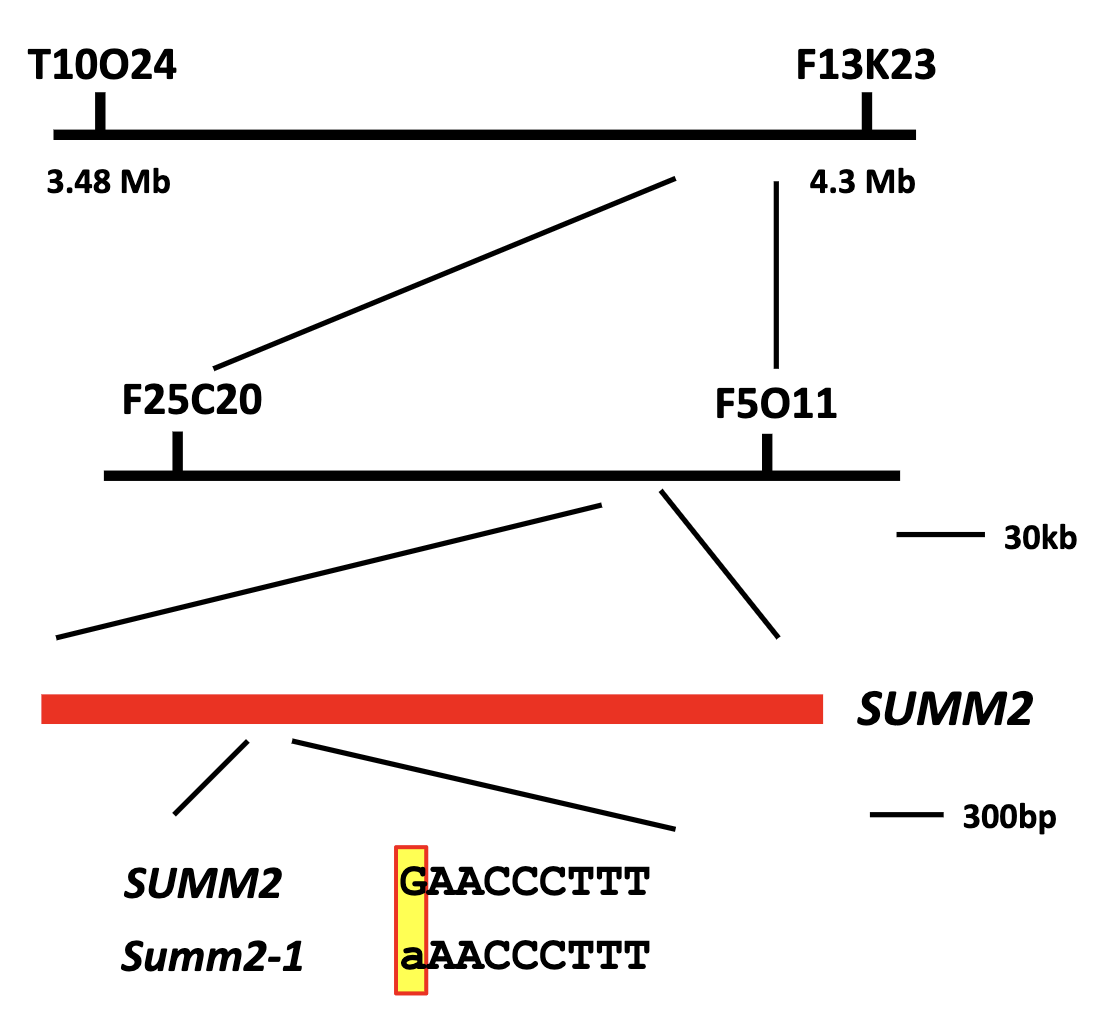
next step: want to clone the summ2 gene; positional cloning
Positional Cloning of summ2
Linkage analysis and fine mapping located gene to a 300 bp region
Sequencing identified a point mutation in one of the genes.
How do they know they cloned the right genes?
other mutants in that same gene have the phenotype
acquired a different lof mutant that has T-DNA (plant transposon) within this suspected gene (strain summ2-8)
this mutant is WT for all the other genes
crossed this mutant w/ the mkk1/2 mutant to see if the summ2-8 mutation could suppress the mkk1/2 phenotype
WT version of summ2 promote apoptosis
How do we know we cloned the right gene? FIGURE
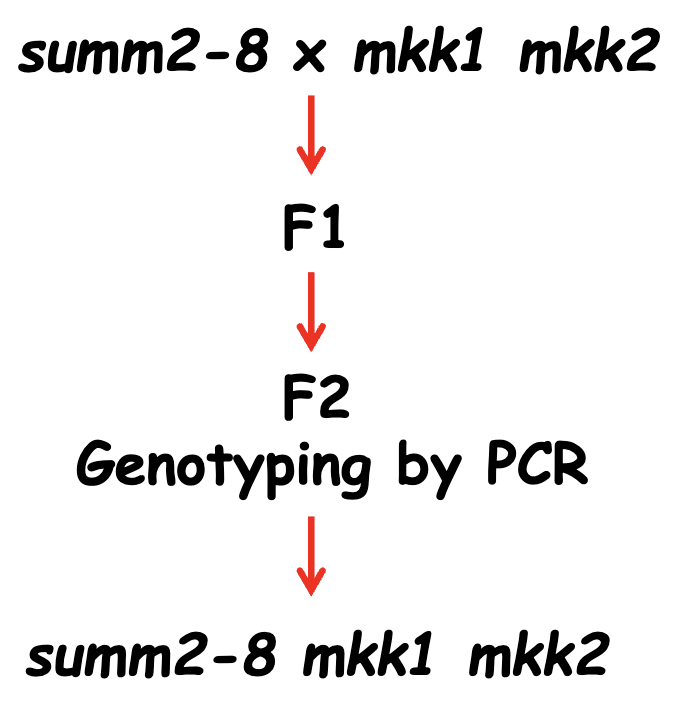
Epistatic Analysis
SUMM2 is epistatic to MKK1/MKK2, which places it downstream of MKK1/MKK2
SUMM2 encodes a protein similar to Apaf1 and Ced4 (which both promote apoptosis)
SUMM2 is episatic to MPK4, which places it downstream of MPK4
SUMM1 is required for cell death in mkk1 mkk2
SUMM2 is epistatic to SUMM1, so it acts downstream of SUMM1
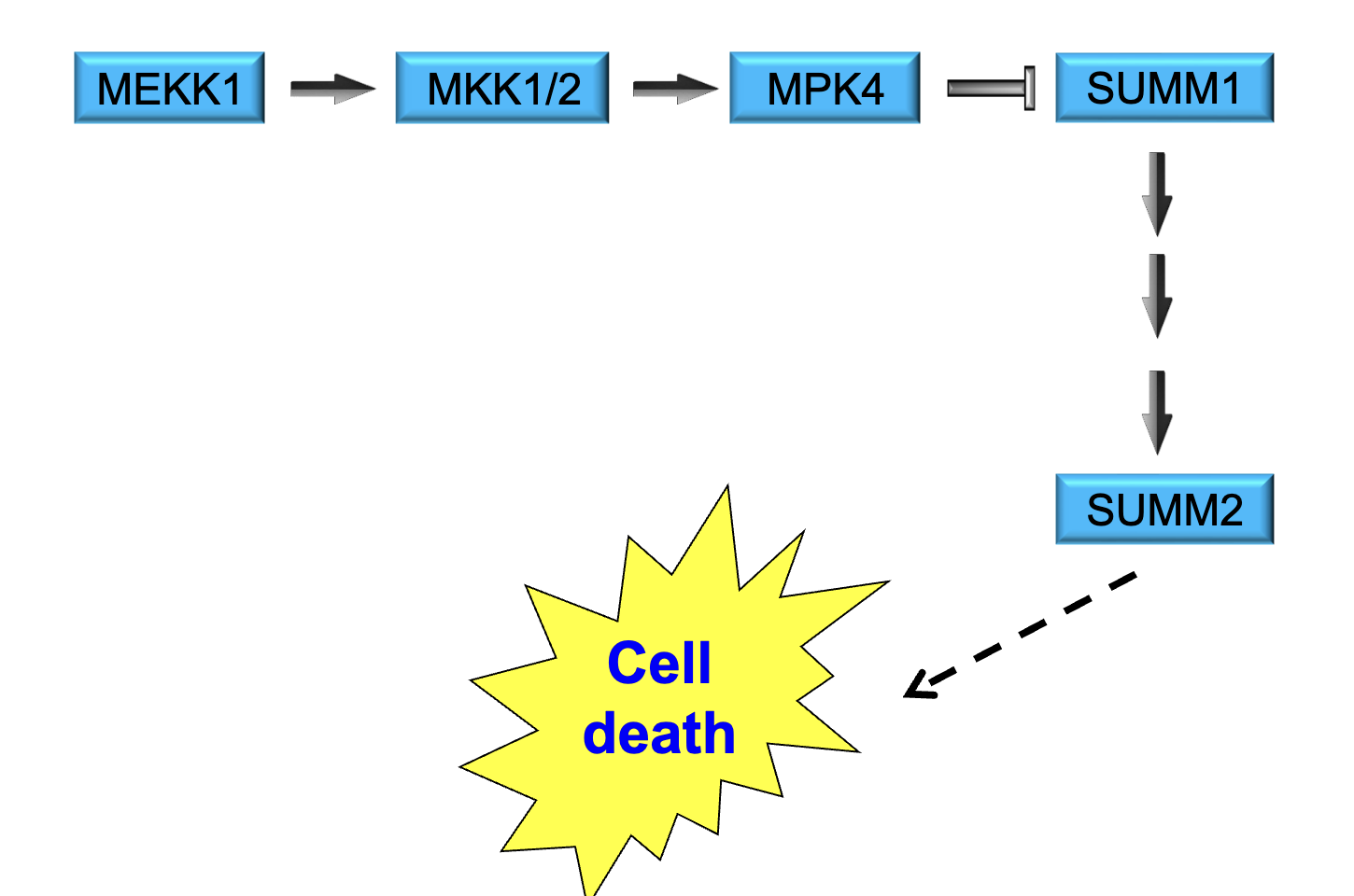
Identification of Cyclin in Sea Urchins
levels of cyclin (protein) cycled regularly around cell division
cyclin levels increased before cell division
cdc Mutants in S.pombe
WT did not grow in restrictive temp
cdc2 mutant grew at restrictive temperature
points to the WT version controlling cell division
Complementation: Genetic Analysis Tool
Two approaches
if we cross 2 mutants w/ the same phenotype, will it reveal that the mutations are in 2 diff genes, or are they mutant alleles of the same gene? (can the mutants complement each other’s mutations in the progeny)
can we restore the WT phenotype if we transform the mutant w/ the cloned gene of interest? (can the clone complement this mutation?)
Human version of cdc gene was found thru complementation experiments (across species)
human gene could complement a mutation in yeast cell
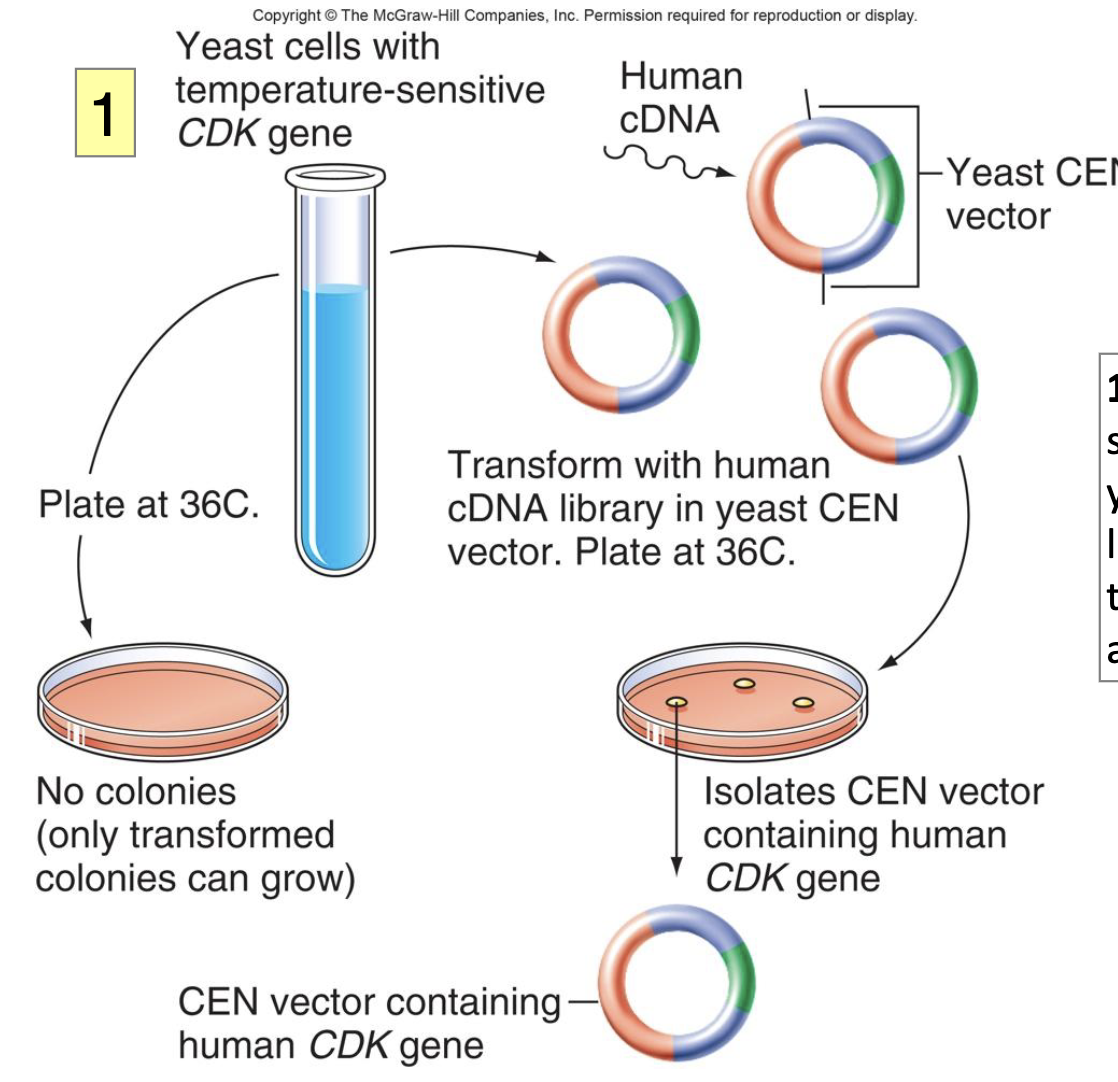
Human cdc Complementation Experiment: Steps
a temp-sensitive allele of the yeast cdk gene that lives at lower temp but dies at 36ºC (protein can no longer fold properly) → will see mutant phenotype
transform yeast mutant w/ a human cDNA library in a yeast CEN vector, where each human gene is expressed from a yeast promoter
the library represents thousands of human transcripts
select clones that are able to survive at 36ºC; these clones have the human cDNA that can rescue the death phenotype
observed: remarkable similarity b/w yeast and human CDC2 protein (abt 63% homology in a.a seq)
similar function since they can substitute for the other
Human cdc Complementation Experiment: Figure
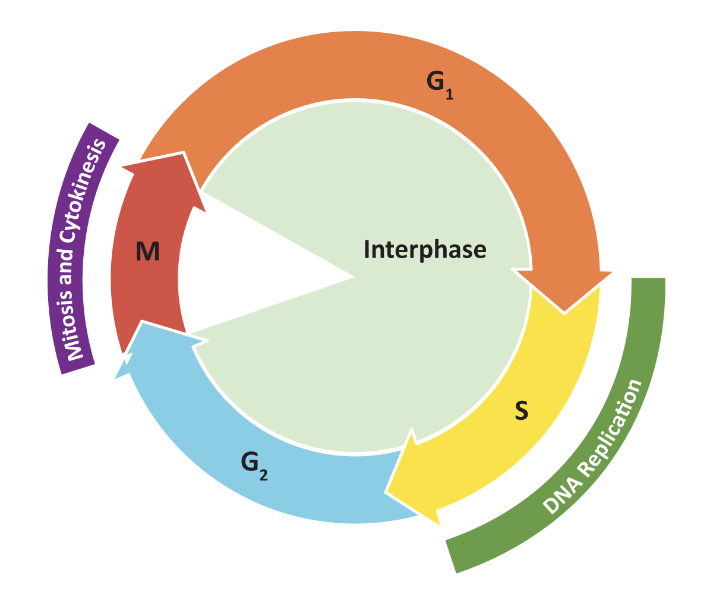
Control of the Cell Cycle: Cyclin/Cdks
Cdc28 (S.cervisiae) and Cdc2 (S. pombe) are cyclin-dependent kinases (i.e Cdk) that regulate entry into mitosis
cyclin/cdks regulate cell cycle progression thru 2 major checkpoints:
G1/S Checkpoint: entry into S phase
G2/M Checkpoint: entry into mitosis
3rd checkpoint to exit mitosis
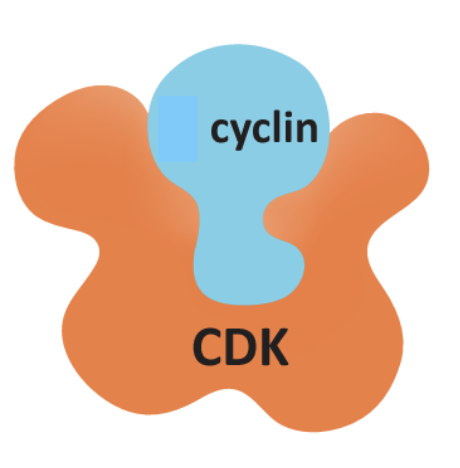
Control of Cdk Enzymatic Activity
Controlled by 2 independent processes:
1) Presence/absence of appropriate cyclin (a regulatory protein): this is influenced by increased expression or targeted degradation of cyclin
2) addition and removal of phosphate groups: phosphorylation of some sites inhibit Cdk activity, others activate
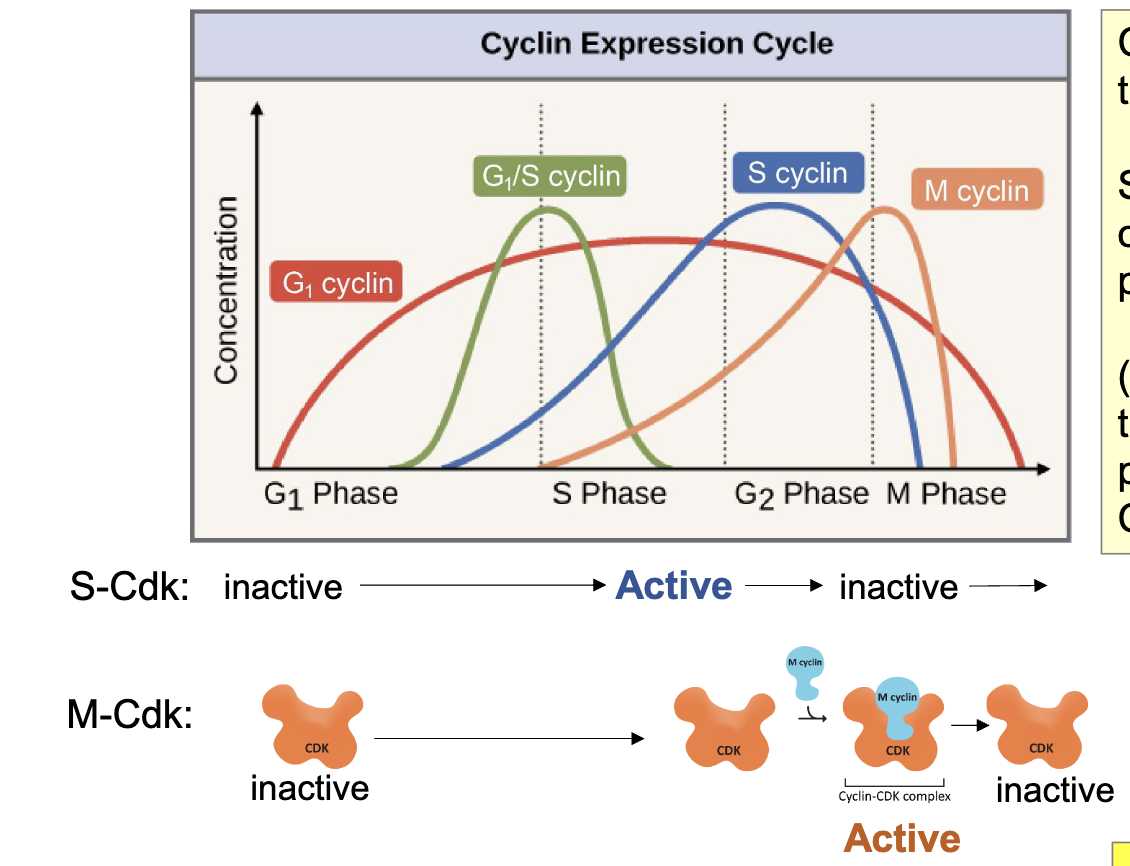
Cyclin-Cdk Pairs Control Entry into Phases
diff cyclin-Cdk pairs control entry into S-phase and into M-phase
Cdks are present throughout cell cycle
specific cyclins are only present at certain point of the cell cycle (additional regulation thru phosphorylation of Cdk)
Cyclin/Cdk Activity in Signal Transduction Pathways
both cyclin and Cdk activity are regulated in signal transduction pathways
expression of cyclins can be induced to promote Cdk activity
Cdks are targets of other kinases and phosphatasese; activity is regulated by phosphorylation/dephosphorylation
Cdk/cyclin complexes are targets of other regulatory proteins, that activate or inhibit Cdk activity thru direct interaction
active Cdks phosphorylate other proteins to promote events in S phase and mitosis
e.g. for DNA replication: Activation of DNA helicase; inactivation of a protein that regulates the origin of replication
e.g for mitosis: activation of Anaphase promoting Complex
Cyclin-Cdks: Cellular Signals
Cyclin-Cdks receive cellular signals and allow the cell cycle to respond to them
cell growth signals (e.g. growth factors): activation of the Cyclin/Cdk complex will allow the cell to proceed to the next phase
growth inhibitory signals (e.g. DNA damage): blocking activation of the cyclin/cdk complex will be maintain the checkpoint ;cell will not proceed
Rb Protein in Mammalian Cells
Rb protein binds to E2F transcription factor and keeps it inactive
a cyclin-Cdk (Active G1-Cdk) appears in late G1 that phosphorylates Rb, inactivating it and lets go of E2F
E2F is now free to transcribe genes important in DNA replication (genes encoding enzymes for replication)
i.e S cyclin and G1/S cyclin
lof mutation in Rb encoding gene would result in failure to arrest in G1 phase
if RB isn’t working then it cannot block E2F activity, and E2F will drive expression of the genes required for S phase.
Modulating Cell # thru Cell Cycle and Apoptosis
both cell division and death can be promoted or inhibited by internal/external signals
the cell cycle is allowed to proceed only if conditions are favorable
if not favorable, the cycle needs to be arrested until the problem is corrected or cell death is induced
Modulating the Cell Cycle: Examples of Regulation
DNA damage inhibits entry to S until DNA repair is repaired
During development, many types of cells require stimulation to divide or undergo apoptosis
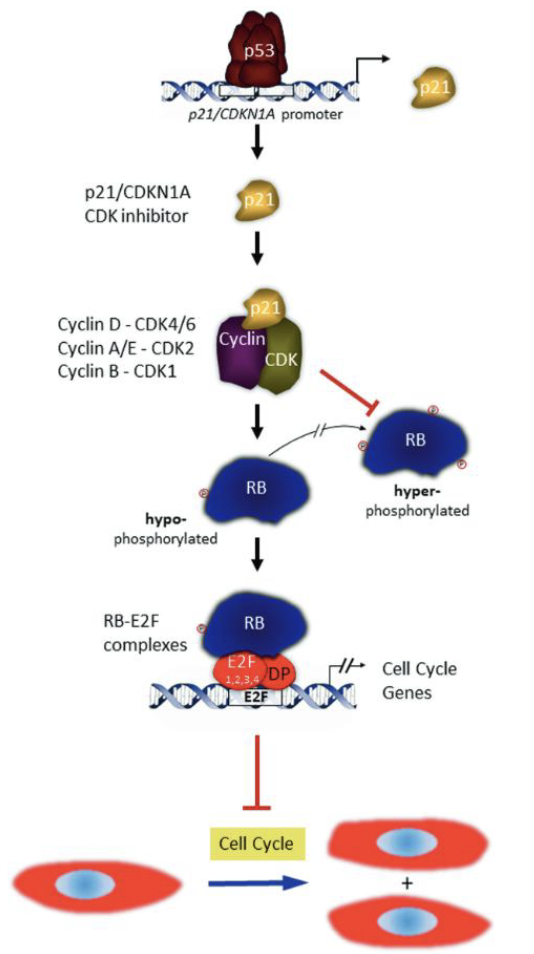
p53, p21 and inhibition of E2F
In mammalian proteins again (Rb protein)
DNA damage causes expression of p53 → p53 turns on expression of p21 → p21 binds to g1 cyclin-cdk complex to stop it from working
Cdk cannot inactivate Rb, Rb continues to hang out to E2F (hypophosphorylated) and prevent steps for DNA synthesis
Rb is a tumor suppressor, a lof in this gene is associated w/ cancer (i.e unregulated cell proliferation)
During development, many types of cells require stimulation to divide or undergo apoptosis
e.g. cell signaling and neuronal development
more neurons are produced than needed
these neurons compete for target-derived neurotrophic factors (e.g. Nerve Growth Factor) and receive survival signals only if they make appropriate connections
e.g. Apoptosis can be induced or repressed by extracellular signals (e.g. FasL system in cytotoxic T lymphocytes)
e.g. growth factor receptor signaling pathways activates cyclinD/Cdk-4/6 which inhibits and promotes E2F and Myc transcription factor
Signaling Proteins are targets for cancer-therapy
generally, signal transduction pathways promote cell survival and signaling
all components of signaling pathway are targets for cancer therapies
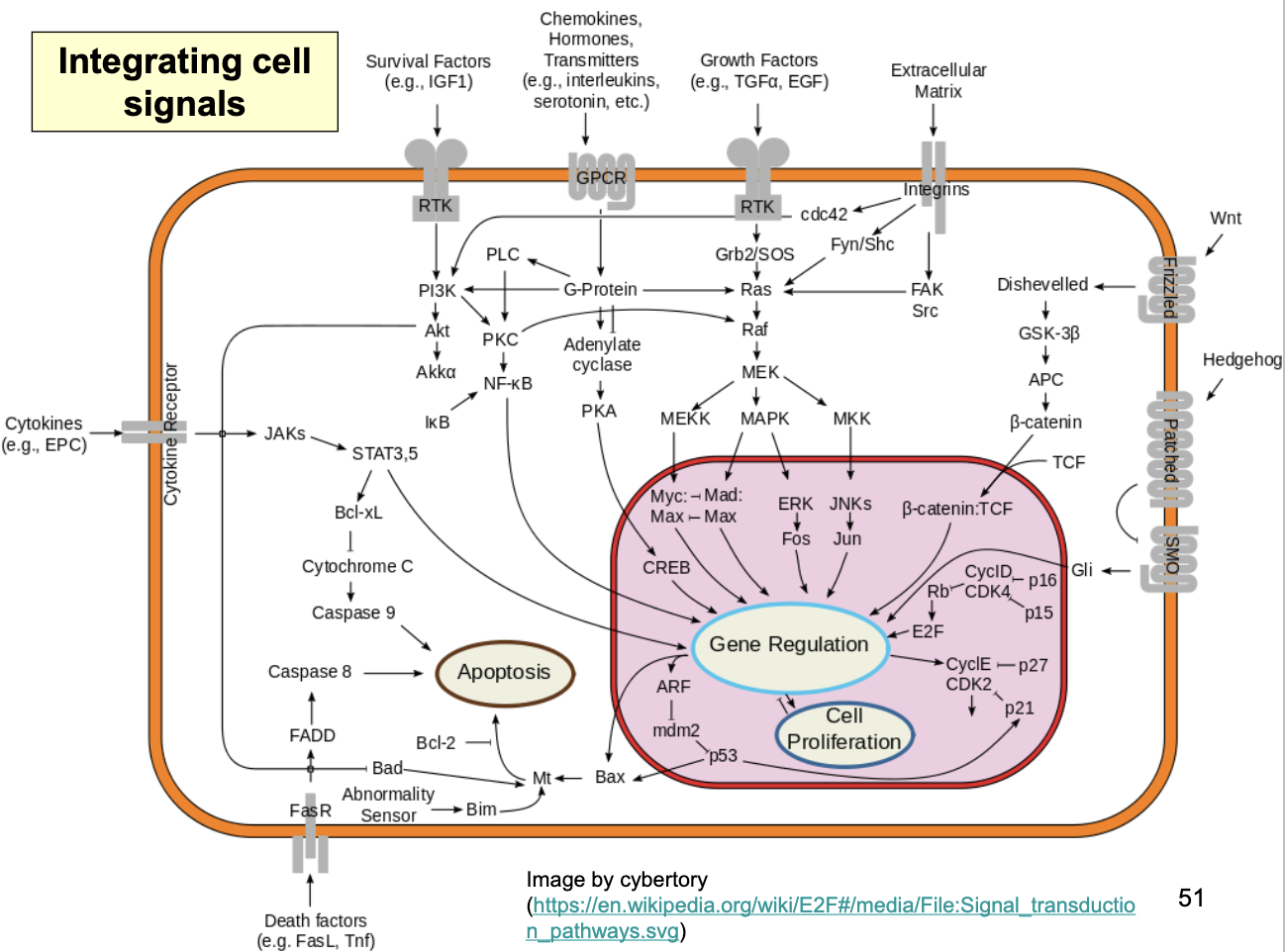
Integrating Cell Signals
Growth factors that act thru Ras/MAPK pathway
survival signals received at the cell surface (survival factors; attachment signals thru integrins) inhibit against apoptosis
signal transduction pathways that regulate Cdk activity
p53 suppresses cell cycle and can promote apoptosis thru Bax (which acts on mitochondria)