Genetics Lab Final Study Guide
1/38
Earn XP
Description and Tags
Exercises 1, 8-14
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
39 Terms
Autosomal Dominant Inheritance
Trait should not skip generations.
An affected person married to a "normal" person should have approximately 50% of the offspring being affected. (Also indicates that the affected individual is heterozygous).
Why not homozygous? Because the person wouldn’t survive past fetal development.
Distribution of the trait should be close to equal among the sexes.
Autosomal Recessive Inheritance
Trait often skips generations.
Traits are often found in pedigrees with consanguineous marriages.
If both of the parents are affected, all of the children should be affected.
Affected individuals can have "normal" parents.
When a "normal" homozygous person is married to an affected individual, all of the children are normal
If a "normal" heterozygous person is married to an affected individual and one or more of the children is affected, then approximately half of the children should be affected.
Sex linked Dominant Inheritance
Trait should not skip generations.
Affected males must come from affected mothers.
Approximately half of the children of an affected female are affected. (Figuring the mother is heterozygous)
All the daughters, but none of the sons, of an affected father are affected.
For a female child to be affected, the father or the mother must be affected (present the mutation).
Sex linked Recessive Inheritance
Most of the affected individuals are males.
For a female child to be affected, the father must be affected and the mother must be affected or a carrier.
All of the sons of an affected mother must be affected.
For a male child to be affected, the mother must be affected or a carrier. (Many times this can be determined by studying males in the mothers family line)
Approximately half of the sons of carrier females should be affected.
Sex linked Inheritance
Most of the affected individuals are males.
If a male child is affected, then the father would also be affected
Process of Chromatography
draw a line in inch from bottom of paper, mark 1-in intervals (baseline)
decapitate flies of same sex and crush 2 heads of same phenotype on appropriate marks for phenotype on paper
bend paper into a cylinder and staple it so edges don’t overlap
place chromatography paper in the chromatograph jar with the baseline down in the solvent (ammonium hydroxide : n-propanol). Make sure the solvent is below the baseline otherwise the pigments will dissolve down into the solvent.
place foil over jar and let the paper develop in the solvent until it’s within 1 inch of the top (will take about 2-4 hours)
Remove staples, place paper in drawer to dry
place under UV light to visualize

Chromatograph Analysis
Calculating Rf
3 results of mutations in metabolic pathways
Rf = (pigment distance) / (total solvent distance)
Compare the mutants to wild type and look for missing precursors and precursors which have accumulated. For some strains, you may be limited in the conclusions you can reach, since not all pigments are visible on the chromatograph.
Consider the three results of mutations in metabolic pathways:
Lack of end product in the pathway
A dysfunctional enzyme in the pathway prevents the pathway from being completed and therefore the synthesis of the final product
Buildup of metabolic precursors
A dysfunctional enzyme prevents the pathway from continuing, while simultaneously causing the buildup of an intermediate molecule due to the enzyme being unable to break it down.
Activation of alternate pathways to neutralize chemical intermediates.
Doesn’t always happen, but often a cell is able to use other enzymes to neutralize an accumulating chemical intermediate. This can be accomplished by “shunting” the molecule into an alternative pathway
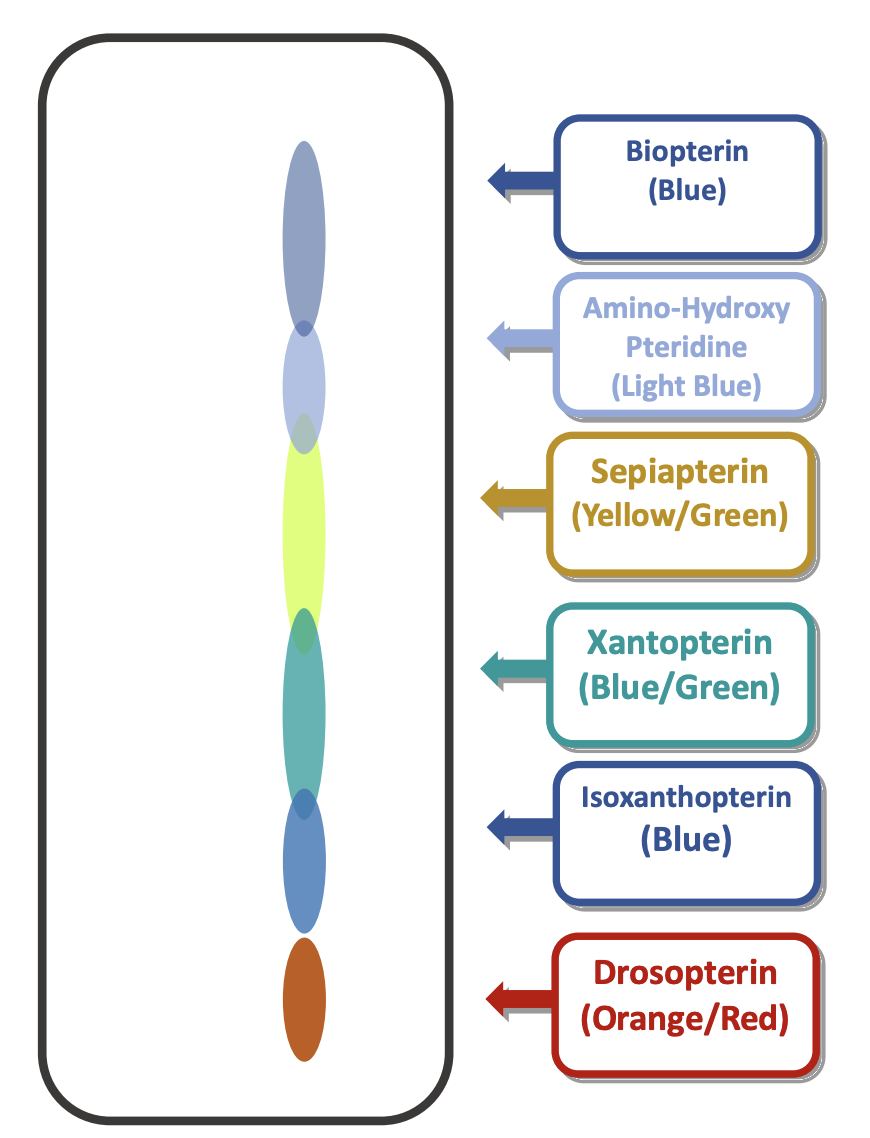
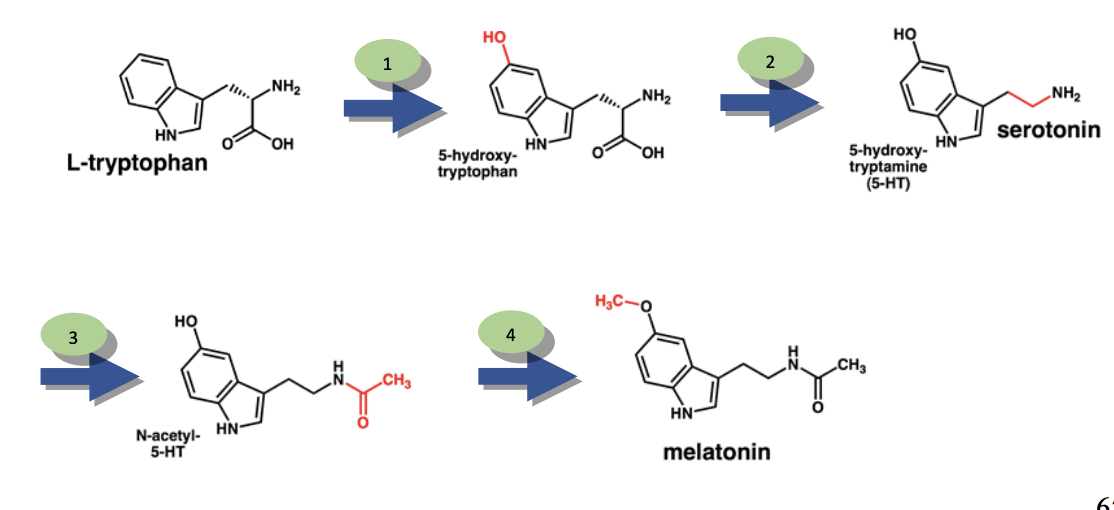
What does it mean when there is an abundance of a specific product on the chromatograph?
using image for an example, let’s say that there was an abundance of 5-hydroxy-tryptophan on the chromatograph
in the example provided in the image, the cause would be a dysfunctional second enzyme.
in this case, the first step of the pathway would occur, producing 5-hydroxy-tryptophan, however, the dysfunctional second enzyme would cause a buildup of that product since it can’t move any further in the pathway.
this type of problem can cause physiological problems in organisms if the intermediate product is toxic.
What does it mean when a certain product is missing from a Chromatograph?
there is a dysfunctional enzyme that is preventing the completion of the metabolic pathway, causing the final product to be absent
Given a certain phenotypic outcome, can you determine what the likely genotypes of the organism?
epistasis problems!!!!!!!
check D2L
genetics lecture: problem set 4 and 5
Problems based on the Hardy-Weinberg equation
p² + 2pq + q² = 1
p² = frequency of homozygous dominant genotype
p = frequency of dominant allele
q² = frequency of homozygous recessive genotype
q = frequency of recessive allele
2pq = frequency of heterozygote phenotype
The 5 assumptions of the Hardy-Weinberg Equation
The population size is very large. Small populations cause distortions due to a limited pool of alleles.
Mating is entirely random. This assumes that neither of the alleles in question affects mate preference.
Mutation rates are very low. If there is a great degree of mutation in the population which could create new alleles of the gene you are studying, this will throw off your predictions.
Natural selection is not occurring. Ie, that alleles of your gene (or another linked gene) do not affect reproductive success. Otherwise, this will skew your results toward one allele over the other.
There is no migration leading to an influx of new alleles.
What are the steps involved in the Restriction Enzyme Digest?
remove 10 uL of sample from the λ-A and place it in the tube labeled λ-A/B. There should now be 10 uL of λ-A DNA left in original tube
add 6 uL ddH2O, 2 uL of 10X enzyme buffer, and 2 uL of enzyme EcoRI in order to the tube marked λ-A/B. If enzyme (very pH sensitive) is added to the water before the buffer, it will be permanently denatured.
mix solution by flicking or pipetting up and down.
Place the λ-A/B tube in the 37°C water bath for 30 minutes. The enzyme should cut the DNA if you made the recipe properly. While you are waiting, make an agarose gel to analyze the sample when the digest is complete
Remove λ-A/B from the bath
add 2 uL loading dye to λ-A and 4 uL loading dye to λ-A/B, then load the total volume and 10 uL of DNA ladder into 3 successive lanes in the gel. Run the gel until the fragments have sufficiently separated and take a photo on the UV imager.
What are the reagents involved in a Restriction Enzyme Digest and their functions?
6 uL ddH2O: is to bring the reaction mixture to the desired total volume while ensuring the purity of the reaction environment.
2 uL of 10X enzyme buffer: provides an optimal chemical environment for the restriction enzyme to function efficiently
2 uL of EcoRI enzyme: makes double-stranded breaks at a specific recognition site: GAATTC
How does DNA fingerprinting (PCR technique) fit in with the AFLP fingerprinting?
Selective amplification: AFLP starts with a complete digestion of genomic DNA, but the goal is to not amplify all fragments. Instead, primers are designed to have a "selective" extension at their 3' end, meaning they only amplify fragments that have specific nucleotides at the ends of the restriction sites. This results in a manageable number of amplified fragments, or "fingerprints".
Creating the fingerprint: The PCR process is repeated many times in a thermal cycler to make many copies of these selected fragments. These amplified fragments are then separated by size using gel electrophoresis and visualized, creating a unique pattern of bands that serves as the DNA fingerprint.
Enabling fingerprinting of any organism: The PCR-based approach allows AFLP to be applied to any organism without prior knowledge of its DNA sequence, making it a versatile tool for DNA fingerprinting.
What are the steps of the PCR?
Using your spectrophotometer readings (given in μg/µL), determine the concentration of your sample in ng/uL (convert from ug/µL if necessary). → (needed DNA conc ng/mL x total volume uL) ÷ (original DNA conc)
pipet 3 uL of your gDNA sample in each 200 uL PCR tube and ddH2O into the control tube.
Make the following PCR master mix in a 1.5 mL tube, enough for five samples (third column). This creates enough PCR solution for your two gDNA samples, + control and - control; always do one more than you have to negate pipetting errors):
Make sure this solution is well mixed by inverting/flicking the tube several times. The glycerol in the Taq is especially dense and will not mix thoroughly unless you forcibly distribute it.
Spin the reaction tubes briefly (about 10 seconds) at low speed (~3000 rpm) in the centrifuge to collect the solution at the bottom of the tube.
Pipet 17 uL of the master mix into each sample tube that already contains your 3 uL of gDNA.
Flick the reaction tubes a few times to distribute the mixture (this helps combine the DNA with the master mix).
Place the reaction tubes in the thermal cycler and initiate the “ITS” program
The reaction itself will take several hours, after which time the instructor will store the samples, to be analyzed next week.
How do you design a primer for pre-amplification and selective PCR?
the primer is complementary to the adapter. Adding an extra nucleotide to the primer prevents some of the fragments from being amplified.
This allows for only ¼ of the fragments to be amplified.
the reverse primer adds 1 extra nucleotide increasing specificity and decreasing the number of amplified fragments to ¼
In expt: E-ACT and M-CAT used. Each has an added 3 selective nucleotides.
What are the steps in AFLP DNA Fingerprinting?
Restriction Digest
Adapter Ligation
Pre-amplification PCR
Selective PCR
Polyacrylamide Gel Electrophoresis (PAGE)
What is the purpose of PAGE in AFLP?
to detect the fragments created by the AFLP process, we need to run the AFLP reactions on an acrylamide gel, which has a greater ability to separate small fragments than do agarose gels
This is important because many of the fragments created by AFLP differ in length by only a few nucleotides.
We will be running a large acrylamide gel identical to those used for DNA sequencing.
How does the acrylamide : bisacrylamide ratio determine the separation of DNA?
the speed of macros to move through an acrylamide gel is dependent on the conc. of an acrylamide and the ratio of acrylamide : bisacrylamide
24:1 is the loosest mesh providing the highest migration, lower resolution
19:1 standard ratio, mid range migration
10:1 tightest mesh, lower migration, higher resolution
What are the types of buffers used for DNA gels
TAE (Tris-acetate-EDTA)
TBE (Tris-borate-EDTA)
TPE (Tris-phosphate-EDTA)
What are the ways that DNA bands can be visualized using PAGE?
2 basic methods
Labeling the DNA fragments with a 32P isotope and place the gel on a piece of X-ray film. This is hazardous due to radiation exposure.
Using silver nitrate to stain the DNA fragments. The chemical basis of this procedure is that silver ions (positively charged) will bind to the negatively charged DNA. The addition of a reducing agent then converts these silver ions into a solid precipitate which shows up as a dark band in the gel
What are the chemicals and their roles used for the silver staining procedure?
10% acetic acid: Removes electrophoresis buffer from the gel and prevents DNA from diffusing out of gel.
ddH2O: First removes acetic acid from the gel and neutralizes pH. Then removes excess silver stain from the gel to prevent entire gel from turning black.
Silver Stain (0.1% silver nitrate, 0.06% formaldehyde): silver ions are attracted to the (-) charged phosphates of the DNA and bind to it
Developer Solution (0.28M sodium carbonate, 0.07% formaldehyde, .0025 mg/mL sodium thiosulfate):
Formaldehyde reduces the silver ions to an insoluble precipitate (black).
Sodium thiosulfate: dissolves insoluble, unbound silver ions from the gel surface to reduce “background” precipitation of silver unbound to the DNA.
What is the procedure for Silver Staining?
Soaking the gel in FIX/STOP solution (10% acetic acid). This removes electrophoresis buffer from the gel and prevents DNA from diffusing out of the gel.
Rinsing with water. Remove acetic acid from the gel and neutralizes the pH.
Incubating the gel in SILVER STAIN (0.1% silver nitrate, 0.06% formaldehyde) The silver ions are attracted to the negatively charged phosphates of the DNA and bind to it.
Another water rinse. Removes excess silver stain from the gel (otherwise the whole gel would stain black. 5)Incubation in DEVELOPER solution (0.28M sodium carbonate, 0.07% formaldehyde, .0025 mg/mL sodium thiosulfate). The formaldehyde reduces the silver ions to an insoluble precipitate (black color). Sodium thiosulfate dissolves insoluble, unbound silver ions from the gel surface to reduce “background” precipitation of silver unbound to the DNA. 91
NOTE: It’s very important to have ultra-pure water with no solutes in it so that other ions do not interfere with the reduction reaction.
How do you calculate the % polymorphism and interpret the data?
% polymorphism = (# of different bands) / (total # of bands) x 100
% polymorphism is the % of difference between samples
e.g. 75% polymorphism means there is a 75% difference btw samples
Want lowest % polymorphism -> determines plant identity
Transition
the conversion of a purine to another purine or a pyrimidine to another pyrimidine
purines: adenine and guanine
pyrimidines: cytosine, thymine, uracil
Transversion
conversion of a purine to a pyrimidine or a pyrimidine to a purine
purines: adenine and guanine
pyrimidines: cytosine, thymine, uracil
Principle of DNA sequencing
To determine the order of the four nucleotide bases (A, T, C, G) in a DNA strand by breaking the strand into fragments of varying lengths and then identifying which base terminates each fragment.
These fragments are then separated by size using gel electrophoresis or capillary electrophoresis, and a detector reads a signal (like a fluorescent tag) corresponding to the final base of each fragment, allowing scientists to reconstruct the full sequence from shortest to longest.
What are the roles of the chemicals used in the DNA sequencing process?
DNA template: provides binding sites for primers -> if too much, non-specific amplification
Buffer: keeps solution at a neutral pH so that Taq polymerase activity is unaffected by pH changes. Also aids in polymerization rxn.
dNTPs: substrate for Taq Pol and are building blocks of the replicated strands. Sufficient quantity is req for rxn to succeed. Too much dNTPs will bind to MgCl2 and prevent rxn from proceeding, conc is important.
primer: either forward or reverse, but only need one.
ddNTPs: to cause chain termination
Mg2+: cofactor for polymerization rxn, it stabilizes the nucleotide tri-phosphates. w/out it the rxn won’t proceed. too much = nonspecific binding
ddH2O: acts as the diluent for the components of the rxn. Highly purified H2O is necessary to prevent contamination. Also provides volume needed.
DNA pol: does amplification
How do ddNTPs (dideoxyribonucleotides) work?
they are missing an OH group at both the 2’ carbon and the 3’ carbon in the ribose sugar. Because it lacks a 3’-OH group, when a ddNTP is incorporated into a DNA molecule, replication cannot continue.
By placing a small concentration of ddNTP (remember that this is ddATP, ddTTP, ddCTP and ddGTP) in the reaction, we introduce the slight chance of a ddNTP being introduced at any given nucleotide.
As amplification continues, eventually there is a DNA fragment present in the sample that has a ddNTP at every position
Read chromatograph and determine DNA sequence
Each set of fragments produced in this manner will be exactly one nucleotide shorter than the last, so we can run these on a gel and see the nucleotide identity at each position.
In the early days of DNA sequencing, four separate reactions were run, each with a single ddNTP (ex: ddATP) and these were run side by side on a gel and successively shorter fragments were identified.
Sequencing reactions are now run with all four ddNTPs in a single reaction, each of which is labeled with a different colored flourescent molecule.
Advanced DNA sequencers have a laser that detects each terminal ddNTP by color. The color and intensity of the signal are detected by the laser and a computer produces a chromatogram
Steps for Restriction Digestion for AFLP Analysis
Add 20 uL of your 0.05 ug/ul working stock of gDNA sample to a new 1.5 mL tube.
Add 3 uL of 10X EcoRI Restriction Enzyme Buffer to the sample. Note that this dilutes the buffer from 10X to nearly a proper working 1X concentration. Add 5 uL of ddH2O to the sample.
Add 2 uL of EcoRI Restriction Enzyme to the sample. The enzyme is in glycerol and will sink to the bottom, so be sure to mix it well by flicking the tube. IMPORTANT: YOU MUST ADD THE BUFFER BEFORE THE ENZYME. The endonuclease is very sensitive to pH etc. and adding it to unbuffered water will permanently denature it. Do not confuse the tubes containing enzyme and buffer.
Incubate the tubes for three hours at 37 degrees Celsius (optimum growth temperature for E. coli)
After the three hour incubation, the instructor will incubate the samples at 70 degrees Celsius for 15 minutes to permanently denature the enzymes so they cease cutting.
Next amplify the DNA fragments to complete the AFLP procedure.
AFLP PHASE I – RESTRICTION DIGESTION & ADAPTER LIGATION
The AFLP process begins by cutting of the genome with two restriction enzymes, most commonly EcoRI (GAATTC) and MseI (TTAA).
This generally produces fragments that range from 50 bp to 1 kb. In fact, assuming a random distribution of the recognition sites, we can calculate EcoRI to cut about every 5 kb (probability of a specific 6-bp sequence is 46 or every 4096 bp) and MseI to cut about every 256 bp (44 = 256 bp).
Both of these enzymes produce staggered end cuts.
A pair of oligonucleotide adapter molecules, which have staggered ends complementary to the EcoRI and MseI cut sites, are ligated to the fragments with DNA ligase.
This creates DNA fragments that have been digested to a manageable size but have ends of known sequence that we can design primers for.
AFLP PHASE II – PRE-AMPLIFICATION REACTION
Once adapters have been ligated to the fragments, we amplify the DNA using PCR. Just like with RFLP, if we were to amplify the entire population of fragments, too many would be produced and we would see a smear on a gel.
Instead, we use a primer that matches the adapter sequence and cut site, but with a single added base; we generally use A for the EcoRI end and C for the MseI end.
In essence we are amplifying all fragments with an EcoRI cut site that has an adjacent A and an MseI cut site that has an adjacent C. If the probability of these 86 two occurrences is random, this would be 1/16 of the total fragments in the population (chance of a given nucleotide at each site is ¼).
This pre-amplification reaction is designed to reduce the total number of fragments we analyze.
AFLP PHASE III – SELECTIVE AMPLIFICATION REACTION
Even the fragments produced by the pre-amplification reaction would be too numerous to analyze. We therefore perform an additional reaction by diluting the pre-amplification product and using it as template for a second PCR.
In this reaction we use primers that have three selective nucleotides after the cut site to reduce the fragments analyzed to a manageable number. This selective-amplification PCR generally produces between 100-150 different DNA fragments per primer set.
Because of the first amplification reaction, we are restricted to selective primers that begin with A at the EcoRI site and C at the MseI site. This leaves us two selective nucleotides at each site or sixteen possible primers we could choose.
For example, we could amplify our fragments with the primers EcoRI-AGG and MseI-CTT. In total, there are 256 possible combinations we could choose which would produce an estimated 25600 unique DNA fragments from the genome.
Note that these fragments are random sequences of DNA scattered throughout the genome. This random nature makes AFLP very useful for determining evolutionary relationships between species because you are essentially screen the whole genome for differences.
However, it makes this technique less useful for mapping specific chromosomes or regions.
AFLP PHASE IV – POLYACRYLAMIDE GEL ELECTROPHORESIS
In order to detect the fragments created by the AFLP process, we need to run the AFLP reactions on an acrylamide gel, which has a greater ability to separate small fragments than do agarose gels.
This is important because many of the fragments created by AFLP differ in length by only a few nucleotides. We will be running a large acrylamide gel identical to those used for DNA sequencing.
AFLP SELECTIVE AMPLIFICATION
Last week we began the AFLP procedure by digesting the genomic DNA of our plants with the enzyme EcoRI. To save time, the instructor has performed the second digestion with the MseI enzyme, ligated the adapters and performed the pre-amplification reaction.
Today we will be performing the selective amplification reaction so that next week we can run these samples on the sequencing gel and obtain the DNA fingerprint of our species.
We will run three separate reactions on each sample, each with a different combination of selective primers.
Reagents for the Selective Amplification PCR
ddH2O: to create the total needed volume
10X PCR Buffer: to maintain pH for the polymerase enzyme
MgCl2: cofactor for polymerization rxn and stabilizes dNTPs
EcoRI Primer E-ACT: creates boundaries for the intended product
MseI Primer M-CAT: creates boundaries for the intended product
dNTP: substrate for Taq Pol and building blocks for replicated strands
Taq polymerase: does the amplification
Chromatograph interpretation
Because of the natural tendency to conserve purine:pyrimidine pairing (Pu:Pu or Py:Py is very chemically unfavorable), transitions are the more frequent mutations, occurring 10-20 times more 10 20 30 40 50 60 96 frequently than transversions.
Higher rates of transversions in a sequence are often considered evidence of greater evolutionary distance between species.
Transition/ transversion ratios are often used in calculation of phylogenetic trees.
Coding sequences are obviously less prone to mutation, because they are subject to natural selection (a mutation that causes a critical gene to cease functioning will not be seen in the population).
The most common types of mutations seen in coding sequences are degenerate mutations, which change the codon but not the amino acid it codes for. This is one of the reasons we used ITS (a non-coding intergenic spacer region) for our analysis.
Pedigree Analysis Questions
Do two individuals without the trait have children who are affected?
Yes = trait is recessive
Is there sex bias?
Yes = trait is X-linked
Do affected males pass on the trait to all daughters, but not sons?
Yes = trait is X-linked and dominant
if all NO → autosomal