Molecular Biology/ Genetics
1/206
Earn XP
Description and Tags
1hr... check DNA structure
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
207 Terms
Genetics
Study of genes, heredity, and genetic variation in living organisms
Explores how traits are passed from parents to offspring
Examines how genetic information influences characteristics, including physical appearance and disease susceptibility
Classical Genetics:
Based on laws of inheritance from Gregor Mendel (1800s)
Modern Genetics:
Studies how genes pass information using molecular chemistry (1950–present)
Genetics Timeline
1865: Mendel’s laws of inheritance
1900: Rediscovery of Mendel’s work
1944: DNA identified as molecule behind inheritance
1953: Watson & Crick describe double helix structure of DNA
1966: Genetic code determined
1972: Cohen & Boyer develop recombinant DNA technology
1974: Belmont Report issued on use of human subjects in research
1977: DNA sequencing methods developed
1982: GenBank database established
Mitosis - Cell Division
Cell replicates chromosomes and segregates them to produce two identical nuclei, preparing for cell division
Meiosis - Cell Division
Cell division in sexually reproducing organisms that reduces chromosome number in gametes (egg and sperm)
Chromosomes
Long linear strands of DNA packaged with histone proteins
All cells (except gametes) have two copies of each chromosome (homologous chromosomes)
Humans: 2 × 23 chromosomes
22 autosome pairs
1 sex chromosome pair
Karyotype
Chromosomal complement of an individual
Men: 22 autosomes + XY
Women: 22 autosomes + XX
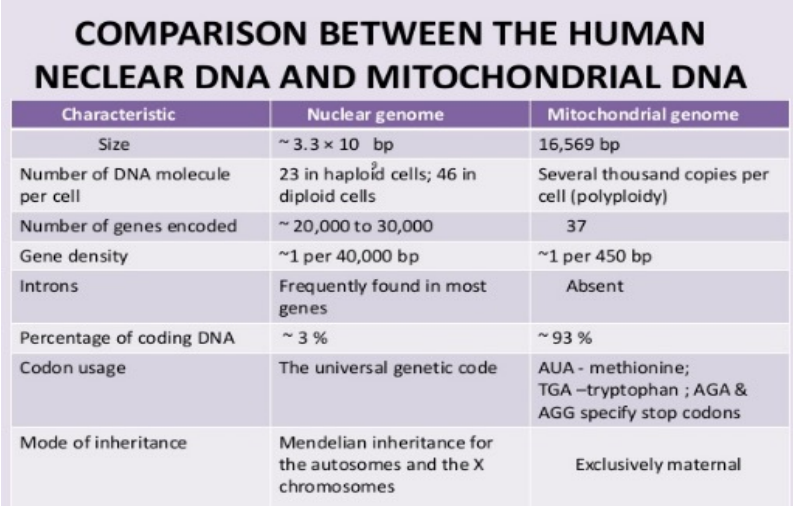
DNA vs RNA
Feature | DNA | RNA |
|---|---|---|
Strands | Double-stranded helix | Single-stranded polynucleotide |
Shape | Stable helix | Can fold into specific shapes |
Location | Nucleus, chloroplast, mitochondrion | Cytoplasm, ribosomes, nucleus |
Function | Stores genetic information | Copies DNA info for protein synthesis |
Sugar | Deoxyribose | Ribose |
Gene
Segment of DNA that contains the instructions for making a specific protein or functional RNA
Responsible for hereditary traits passed from parents to offspring
Can influence physical characteristics, biochemical pathways, and disease susceptibility
Unit of inheritance
Located at a specific locus on a chromosome
Allele
A variant form of a gene at a specific locus on a chromosome
Individuals inherit two alleles for each gene, one from each parent
Alleles can be dominant or recessive, affecting the expression of a trait
Responsible for genetic variation in a population
Homozygote
An individual with two identical alleles for a particular gene at a specific locus
Heterozygote
An individual with two different alleles for a particular gene at a specific locus

Dominant Allele
An allele that expresses its trait even when only one copy is present (heterozygous condition)
Recessive Allele
An allele that expresses its trait only when two copies are present (homozygous condition)
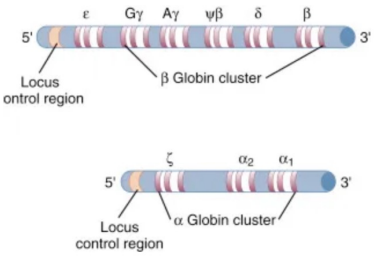
Gene Clusters
Groups of functionally related genes located close together on the same chromosome
Positioned to allow coordinated regulation and controlled expression
Genotype
The genetic make-up of an organism that determines its traits/ phenoytype
Phenotype
The visible characteristics of an organism, resulting from the interaction of genotype and environment
The Human Genome (photo)
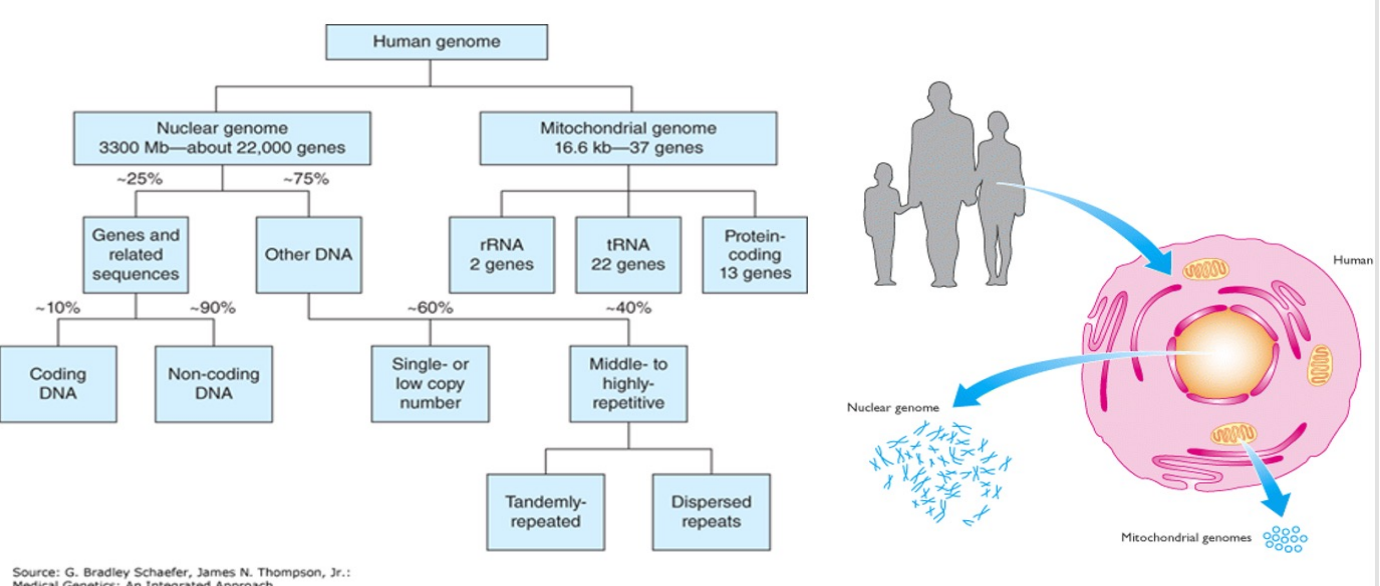
The Human Genome
The nuclear genome provides the great bulk of essential genetic information, most of which specifies polypeptide synthesis on cytoplasmic ribosomes
The mitochondrial genome specifies only a very small portion of the specific mitochondrial functions
The bulk of the mitochondrial polypeptides are encoded by nuclear genes and are synthesized on cytoplasmic ribosomes before being imported into the mitochondria
Nuclear Genome - Human Genome
The nucleus of a human cell typically contains more than 99% of the cellular DNA
DNA is structured in long strands that are wrapped around protein complexes called nucleosomes that consist of proteins - histones
Such structured DNA constitute a chromosome
The human cell has 46 chromosomes:
22 pairs of autosomes
1 pair of sex chromosomes, X and Y
Nuclear genome: 3200 Mb, ~22,000 genes
4.5% highly conserved including 1.5% coding DNA and 3% of conserved untranslated & regulatory sequences
90%-95% of the coding DNA is protein coding while the remaining (5-10%) is untranslated (RNA genes)
The coding sequence is present in families of related sequences generated by gene duplication which resulted in pseudogenes and gene fragments
The 95.5% non-coding DNA of the human genome is made up of tandem repeats (head to tail) or dispersed repeats resulting from retrotransposition of RNA transcripts
Mitochondrial Genome - Human Genome
= mitochondrial DNA (mtDNA)
Is the genetic material found within mitochondria
It's a small circular DNA molecule distinct from the larger nuclear genome located in the cell's nucleus
In humans, it contains 37 genes that code for proteins
16,569 bp
37 genes:
13 code for enzymes of oxidative phosphorylation
2 code for mt rRNAs
22 code for mt tRNAs
Copy Number and Distribution
A single mitochondrion contains 2 to 10 mtDNA copies
A single somatic cell (containing only two chromosome copies) has 100–100,000 mtDNAs
The number of mtDNA can vary considerably in different cell types
Lymphocytes have about 1000 mtDNA
Certain cells, such as terminally differentiated skin cells, lack any mitochondria and so have no mtDNA
Mitochondrial DNA in Gametes
Sperm cells have a few hundred copies of mtDNA
Oocytes have about 100,000 copies, accounting for over 30% of the oocyte DNA
Sperm do not contribute mtDNA to the zygote (strictly maternal)
During mitosis, mitochondria are passed on to daughter cells by random assortment
Mitochondrial Inheritance
During zygote formation, a sperm cell contributes its nuclear genome but not its mitochondrial genome to the egg cell
Mitochondrial genome is maternally inherited: males and females both inherit their mitochondria from their mother
Males do not transmit their mitochondria to subsequent generations
During mitotic cell division, the mtDNA molecules of the dividing cell segregate in a purely random way to the two daughter cells
Nuclear DNA vs. mtDNA (photo)
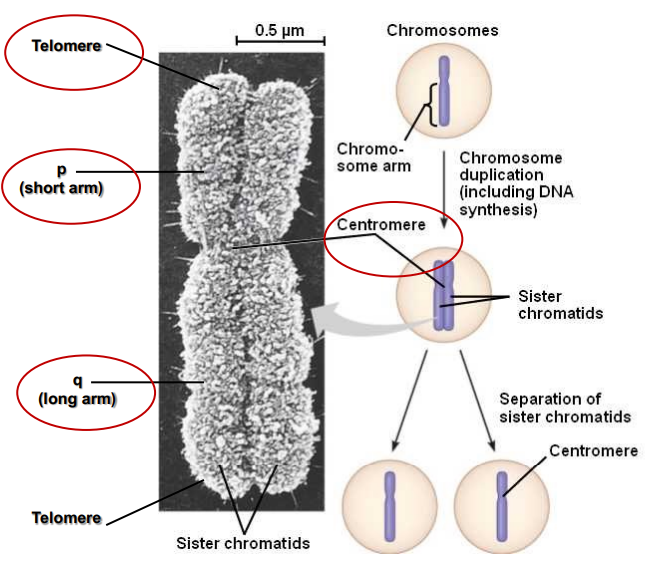
Chromosomes Structure (photo)
Identifying Chromosomes (photo)
G-bands: dark staining bands with Geimsa
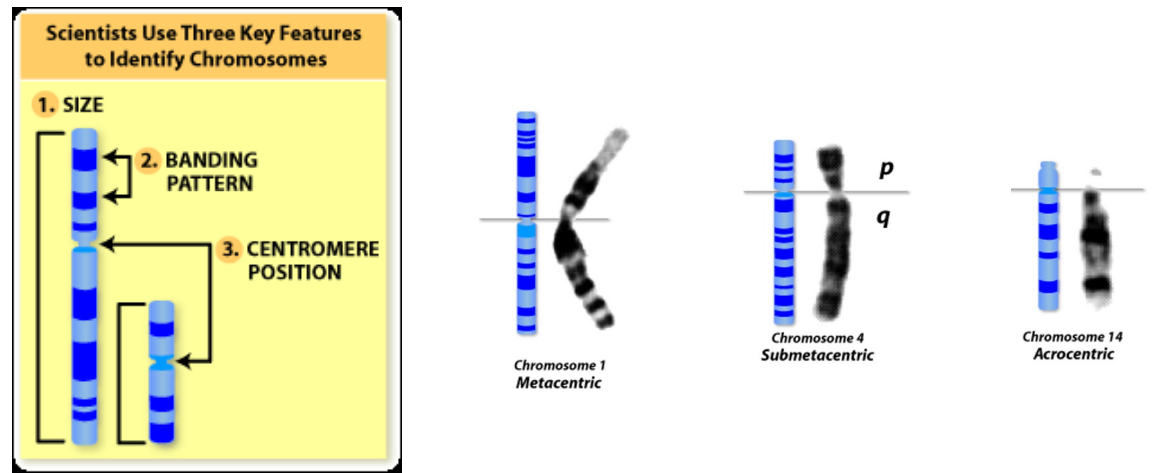
Identifying Chromosomes
Cytogenetic techniques have been used to unravel the three-dimensional organization of the genome and epigenetic features of higher-order chromatin structure
Size of chromosomes
Position of centromere
Chromosomal Abnormalities
A normal human cell contains 23 pairs of chromosomes, including 22 pairs of autosomes and a pair of sex chromosomes (XX or XY)
There are many types of chromosome abnormalities, but they can be organized into two basic groups:
numerical abnormalities
structural abnormalities
Numerical Abnormalities - chromosome
When an individual is missing one of the chromosomes from a pair, the condition is called monosomy
When an individual has more than two chromosomes instead of a pair, the condition is called trisomy
Examples:
Down syndrome (trisomy 21)
Edward’s syndrome (trisomy 18)
Structural Abnormalities (photo)
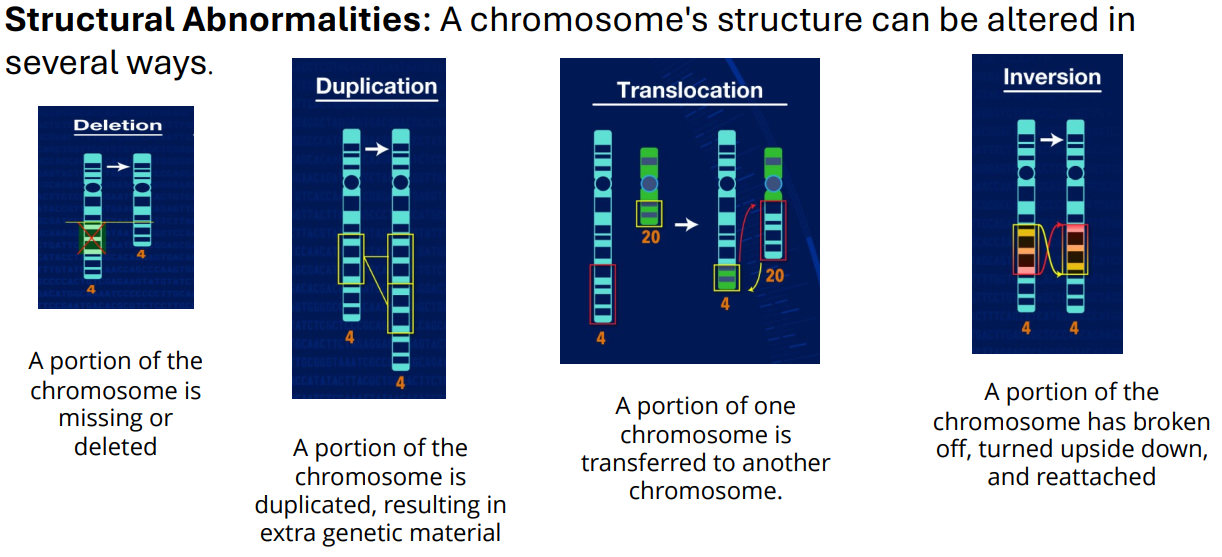
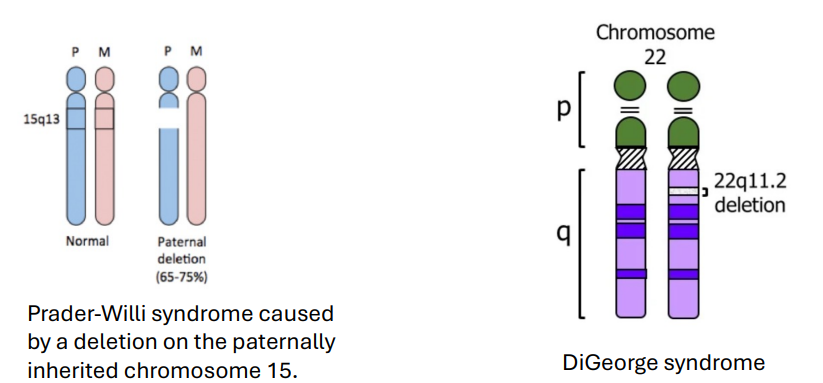
Deletion - Structural Abnormalities (photo)
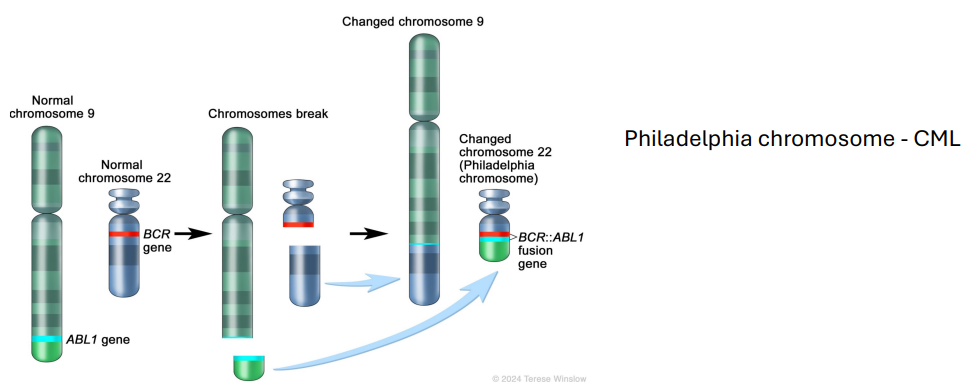
Translocation - Structural Abnormalities (photo)
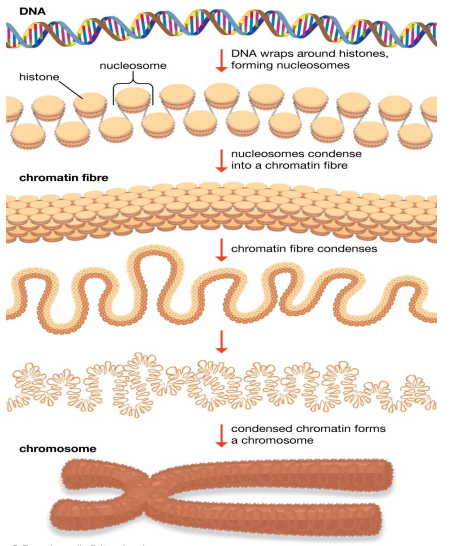
Chromosomes Organisation
Eukaryote DNA is tightly bound to small proteins (histones) that package the DNA in the nucleus
The total extended length of human DNA is nearly 2 meters, but it must fit into a nucleus with a diameter of 5 to 10 μm
Chromatin is a complex of eukaryotic DNA and proteins
DNA to chromosome (photo)
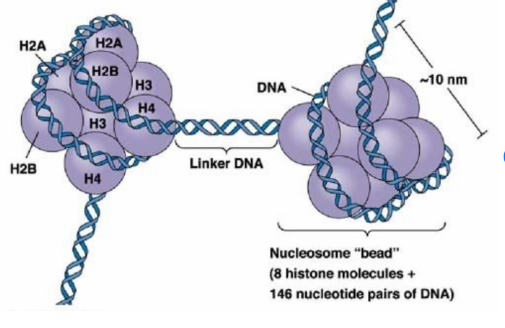
Histones
proteins that package and order DNA into nucleosomes to form chromatin
Small: 10–20 kDa
Highly conserved
Very basic proteins
Heavily acetylated/methylated
A core of 8 histones (2 each of H2A, H2B, H3, and H4) around which the DNA is wrapped
Histone H1 attached to linker DNA between nucleosomes
Do not dissociate from DNA during DNA replication
Histones (photo)
Conformational transition b/n euchromatin & heterochromatin (photo)
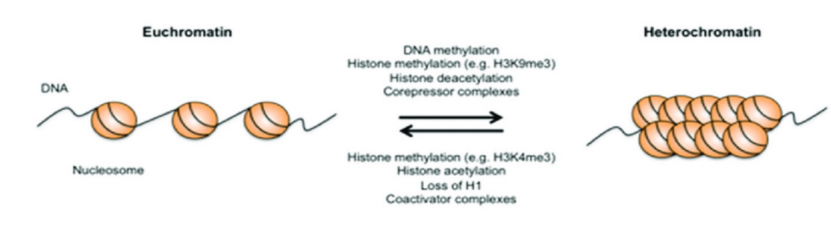
Euchromatin
The fraction of the nuclear genome which contains transcriptionally active DNA and which adopts a relatively extended conformation
Enriched in genes
Often under active transcription
Heterochromatin
A chromosomal region that remains highly condensed throughout the cell cycle and shows little or no evidence of active gene expression
Constitutive: always inactive and condensed (e.g. centromere)
Facultative: can exist as either condensed or dispersed (e.g. mammalian X-chromosome)
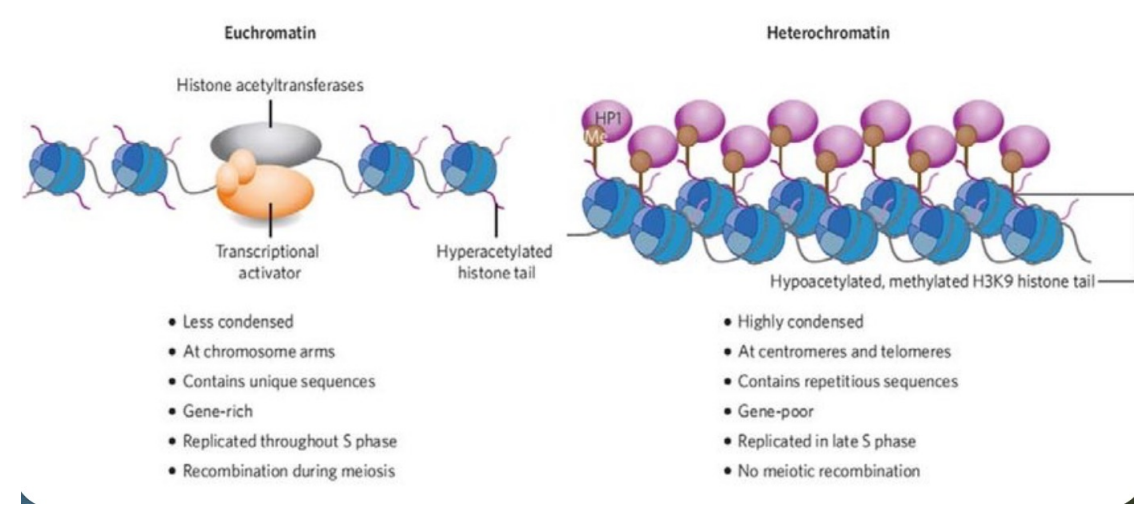
Euchromatin vs. Heterochromatin (photo)
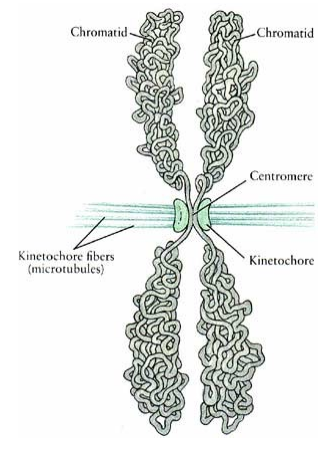
Centromere
Constriction in the chromosome
Region where the sister chromatids are held together
Essential for attachment to the spindle and segregation
A specific DNA sequence which is highly repetitive
Centromeres are DNA sequences to which proteins bind, forming a kinetochore
Centromere (photo)
spindle fibres centre
Telomere
Sequences at the ends of chromosomes required for replication of linear DNA
Repetitive DNA sequence
Protect the ends of chromosomes and prevent loss during DNA replication
Maintain structural integrity of chromosomes
Linked to ageing
Bind a protein complex (shelterin) that protects the chromosome termini from degradation
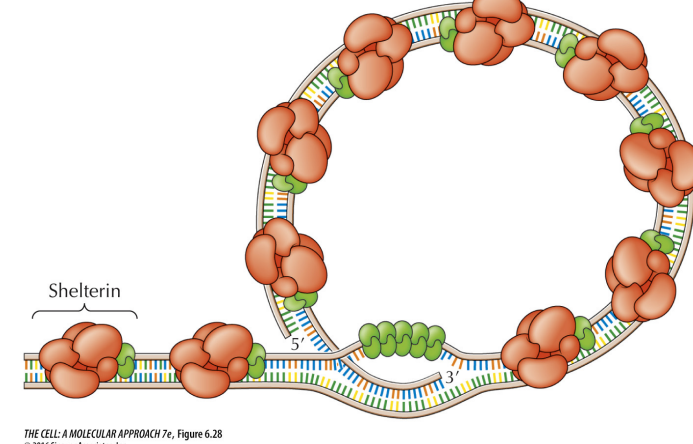
Telomere (photo)
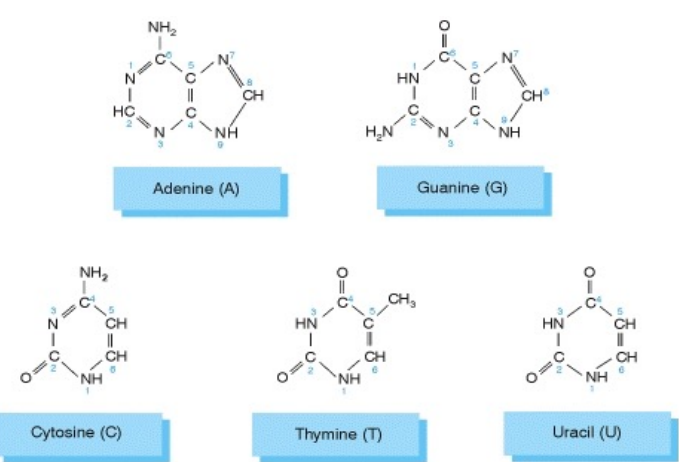
Nucleotides
Linkage Disequilibrium
Unlinked genes are genes located on different chromosomes or far apart on the same chromosome
Unlinked genes are inherited independently of each other
Linkage Group
A set of genes and their alleles located at different loci on the same chromosome
Linked alleles tend to be inherited together
Independent inheritance occurs only when crossing over happens
Penetrance
Measures the proportion of individuals in a population who carry a specific gene and express the associated trait
Mosaicism
A condition in which cells within the same individual have different genetic makeups
Can affect any cell type, including blood cells
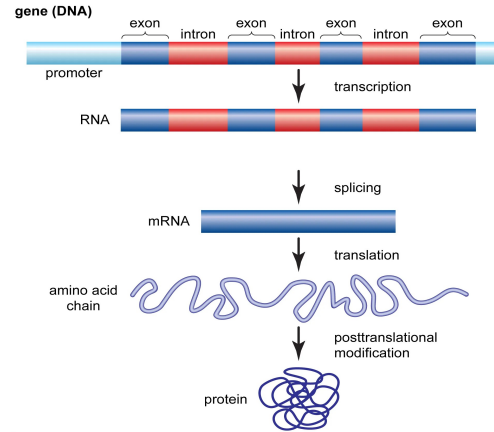
Gene
A segment of DNA that is expressed to produce a functional product, such as rRNA, tRNA, or a polypeptide
Structure of Eukaryotic Genes
Eukaryotic genes contain both coding and noncoding DNA
Noncoding sequences occur both within genes and between genes
Exons and introns
Coding regions called exons are interrupted by noncoding regions called introns
The entire gene is transcribed into RNA
Introns are removed by RNA splicing
Only exons remain in the mature mRNA
Untranslated regions (UTRs)
Exons include regions at both ends of the mRNA
5′ untranslated region 5′ UTR
3′ untranslated region 3′ UTR
UTRs are not translated into protein
Size and structure of human genes
Introns can be much longer than exons
The average human gene contains about 10 exons
The average human gene spans approximately 56,000 base pairs 56 kb
DNA composition
Total exon sequence about 4,300 base pairs
Protein coding sequence about 1,700 base pairs
Intron sequence about 52,000 base pairs
Introns make up more than 90 percent of the average human gene
Gene Structure (photo)
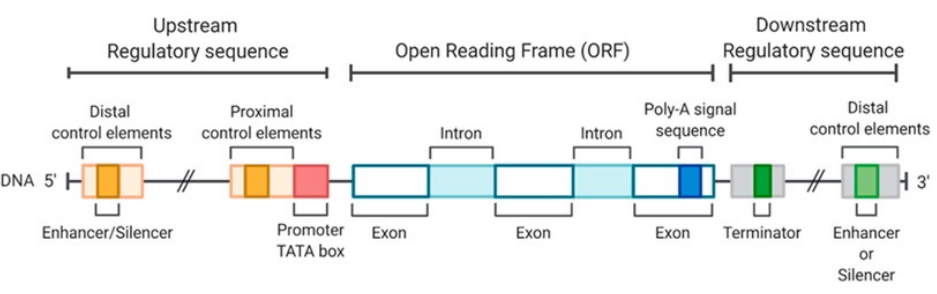
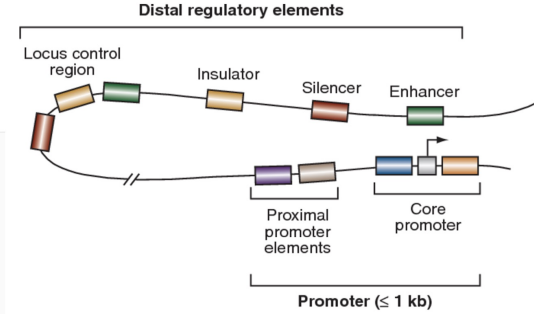
Distal Regulatory Element Structure (photo)
Locus Control Region
Regulatory Elements Involved in Gene Transcription
Distal regulatory elements
Locus control region LCR
Insulator
Enhancer
Silencer
Proximal promoter elements
GC rich box
CAAT box
Core promoter
TATA box
Transcription start site TSS
Distal regulatory elements
Located far from the core promoter
Contain multiple transcription factor binding sites
Can function as enhancers to activate transcription
Can function as silencers to repress transcription
Locus control region LCR
Long range DNA regulatory element
Ensures correct tissue specific expression
Regulates a cluster of linked genes
Functions by altering chromatin accessibility
Insulator
DNA sequence that acts as a boundary or barrier
Controls gene expression by blocking the influence of enhancers or silencers
Prevents inappropriate activation or repression of neighboring genes
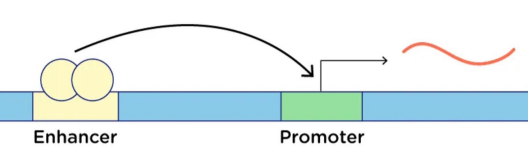
Enhancer
Regulatory DNA sequence that binds transcription factors
Increases the rate of transcription of a gene
Can act at a distance from the target gene
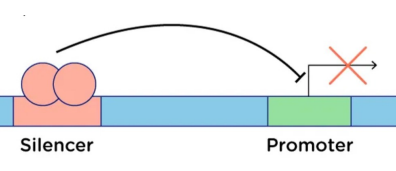
Silencer
Regulatory DNA element that reduces transcription
Acts on its target promoter
Repressive counterpart of enhancers
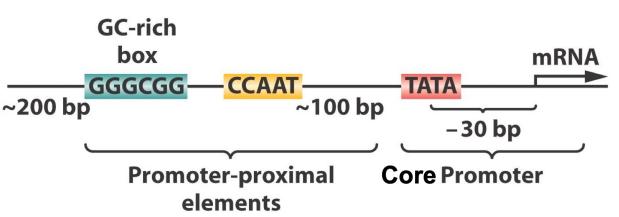
Proximal promoter elements
DNA sequences located close to the transcription start site
Usually within about 200 base pairs of the gene
Bind transcription factors to regulate transcription
Recruit RNA polymerase
Influence how frequently a gene is transcribed into RNA
GC rich box - Proximal promoter elements
Short regulatory DNA sequence rich in guanine and cytosine
Binds specific transcription factors
Enhances gene transcription
CAAT box - Proximal promoter elements
DNA promoter sequence recognized by transcription factors
Stabilizes the transcription initiation complex
Facilitates gene expression
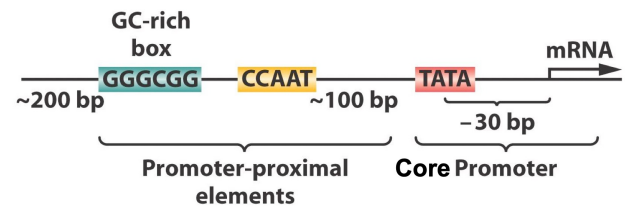
Core promoter
Minimal DNA region required for transcription initiation
Binding site for RNA polymerase and general transcription factors
Essential for the start of gene expression
TATA box - Core promoter
DNA promoter sequence with the consensus sequence TATAAA
Binds the TATA binding protein
Required for initiation of transcription
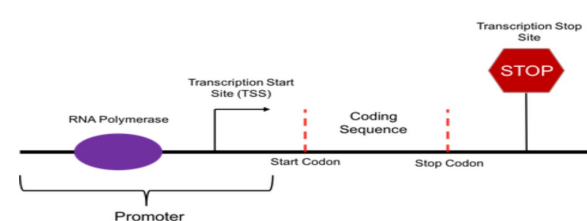
Transcription start site TSS
Specific location on the DNA where transcription begins
Marks the first nucleotide of the RNA transcript
Defines the start of the gene to be transcribed
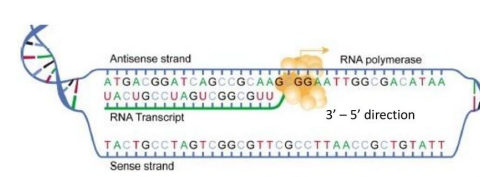
Sense and Antisense DNA Strands
Sense strand
Also called the coding strand
Its sequence determines the protein sequence
Has the same sequence as the mRNA except thymine T is replaced by uracil U
Antisense strand
Serves as the template strand for mRNA synthesis
Complementary to the RNA transcript
Noncoding Sequences in Eukaryotic Genomes
Eukaryotic genomes contain many sequences that do not code for proteins
Many are involved in gene regulation
Some noncoding sequences contribute to chromosome structure and replication
Understanding noncoding sequence function is essential for understanding development and behavior
The ENCODE project analyzed 147 human cell lines to define functions of different sequence types
Approximately 75 % of the human genome is transcribed
This revealed that noncoding RNAs play a major role in gene regulation
Noncoding RNA (ncRNA)
Refers to RNA molecules that do not encode proteins
Lack of protein coding capacity does not mean lack of information or function
ncRNAs have important regulatory and structural roles
Functional RNAs
rRNA
Fundamental structural and functional component of ribosomes
tRNA
Noncoding RNA that acts as an adaptor molecule during protein synthesis
microRNA miRNA
Small noncoding RNA
Regulates gene expression at the post transcriptional level
Silences target messenger RNA mRNA
snRNA
Short noncoding RNA
Essential for mRNA splicing and other RNA processing events
Long Noncoding RNAs lncRNAs
RNA molecules longer than 200 nucleotides
Crucial regulators of gene expression at multiple biological levels
Epigenetic regulation
Transcriptional regulation
Post transcriptional regulation
Function as molecular scaffolds
Interact with DNA, proteins, and other RNAs
Control diverse cellular processes
Dysregulation is implicated in various diseases including cancer
Repetitive DNA in the Genome
Coding regions can contain repetitive DNA sequences
Most highly repetitive DNA is located outside genes
Major categories of repetitive DNA
Heterochromatin
Long arrays of tandem repeats
Located in condensed chromosomal regions
Does not contain genes
Transposon repeats
Interspersed throughout the genome
Account for about 40 percent of the human genome
Found in extragenic regions, introns, and untranslated regions
Heterochromatin vs Euchromatin of Chromosome
Heterochromatin (edge)
More condensed
Genes are silenced and often methylated
Gene poor with high AT content
Stains darker
Euchromatin (mid area)
Less condensed
Transcriptionally active
Gene rich with higher GC content
Stains lighter
Satellite DNA
Constitutive Heterochromatin that are found in:
Centromeres
Telomeres
Most of chromosome Y
Short arms of acrocentric chromosomes 13, 14, 15, 21, 22
Highly condensed and transcriptionally silent
Composed of long arrays of high copy number DNA sequences repeated in tandem
Known as satellite DNA
depending on size The Major types
Alpha satellite
Minisatellite
Microsatellite
Alpha Satellite DNA
Consists of tandem repeats of a 171 base pair repeat unit
Makes up the bulk of centromeric heterochromatin
Present on all chromosomes
Repeat units often contain binding sites for centromere protein CENP B
Plays a critical role in centromere structure and function
Minisatellites
Found as tandem arrays but mostly interspersed throughout the genome
Occur at more than 1000 locations in the human genome
Repeat units typically 10 to 100 base pairs
Also known as VNTRs
Hypervariable minisatellite DNA
Repeat units vary in size but share a common core sequence GGGCAGGAXG where X is any nucleotide
Found mainly near telomeres
Act as hotspots for homologous recombination
Telomeric minisatellite DNA
Located at chromosome ends
Consist of 3 to 20 kb of tandem TTAGGG repeats
Essential for replication of linear chromosome ends
Microsatellites
Mostly found as tandem repeats
Consist of repeat units of 1 to 7 base pairs
Interspersed throughout the genome
Account for over 60 Mb or about 2 percent of the genome
Dinucleotide repeats are the most common
CA TG about 1 per 36 kb
AT TA about 1 per 50 kb
AG TC about 1 per 125 kb
CG GC very rare about 1 per 10 Mb
Mostly located in introns
Rare in exons where they act as mutational hotspots
Example of repeat contraction due to replication errors shown by loss of repeat units
Types of Non Coding Repeats - Repetitive DNA
Minisatellites
Repeat size 10 to 50 bp
Repeated up to 1000 times
Microsatellites
Repeat size 2 to 9 bp
Repeated 10 to 100 times
Applications of Minisatellites and Microsatellites
Used as DNA markers due to high variability
Important in
Forensic DNA analysis
Paternity testing
Implicated in disease
Huntington’s disease
Myotonic dystrophy
Minisatellites and Microsatellites as DNA Markers
Many repeats are hypervariable
Number of repeat copies varies greatly between individuals
Results in many alleles within the population
STR and VNTR analysis widely used in genetic studies
Higher probability of variation compared to non repeating DNA
Variability arises mainly from replication errors
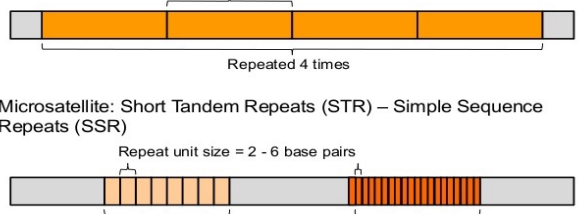
Tandem Repeat Elements - Satellite DNA
Minisatellites
Also called Variable Number Tandem Repeats VNTRs
Repeat unit size in the hundreds of base pairs
Typically repeated a few to many times
Microsatellites
Also called Short Tandem Repeats STRs or Simple Sequence Repeats SSRs
Repeat unit size of 2 to 6 base pairs
Can be repeated from 8 up to 20 or more times
Formation of Tandem Repeats
Replication slippage (also called polymerase stuttering) is the main mechanism
Occurs during DNA replication when DNA polymerase slips on the template strand
Backward slippage
Newly synthesized strand loops out
Results in insertion of repeat units
Forward slippage
Template strand loops out
Results in deletion of repeat units
Thus, replication slippage can increase or decrease the number of tandem repeats, generating variability in repeat length
Mobile Genetic Elements
DNA fragments that can move within the genome
Flanked by short, inverted repeat sequences
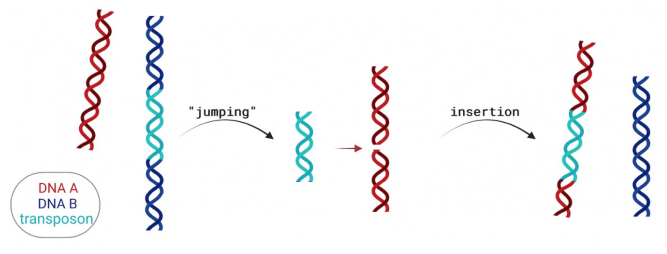
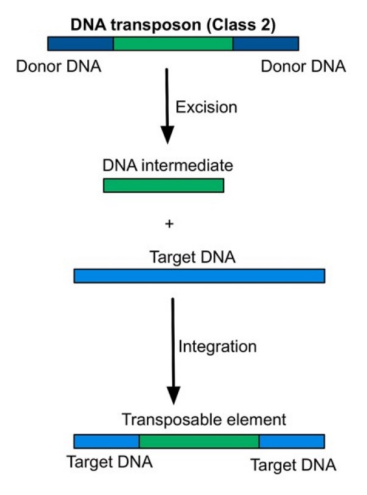
Transposons (DNA Transposable Elements)
Move using a cut-and-paste mechanism
Enzyme involved: Transposase
Mechanism:
Transposase cuts DNA to produce sticky ends
Transposable element is inserted at a new site
DNA ligase fills the gaps
Flanking repeat sequences are recreated at the insertion site
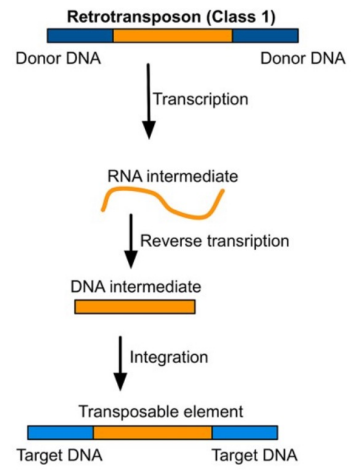
Retrotransposons
Move via an RNA intermediate
Mechanism:
DNA → RNA → DNA
Newly synthesized DNA is inserted into a new genomic location
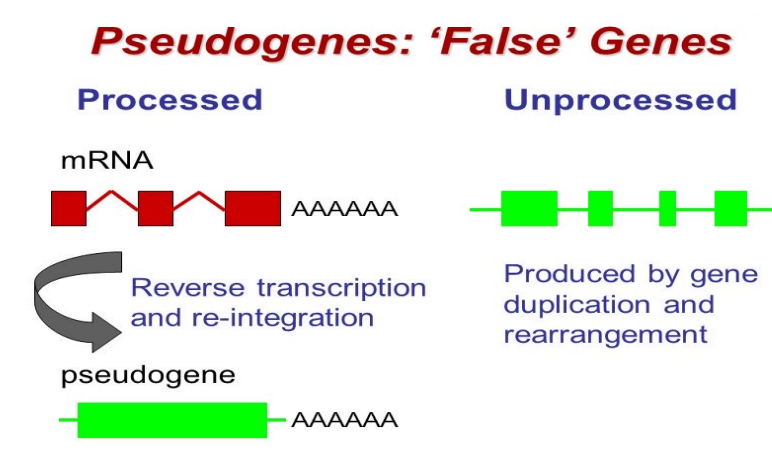
Pseudogenes
Nonfunctional DNA sequences resembling active genes
Inactivated by mutations such as stop codons or frameshifts
Historically considered “junk DNA”
Some pseudogenes are transcribed into functional noncoding RNAs
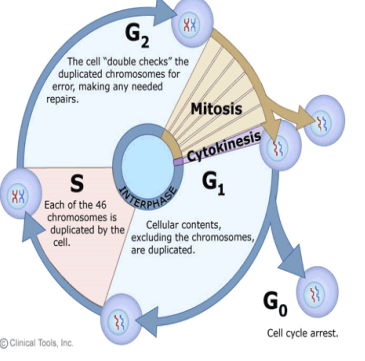
Cell Cycle
Ordered series of events involving cell growth and division producing two daughter cells
Precisely timed and regulated stages of growth, DNA replication, and division
Major phases
Interphase – cell grows and replicates DNA
Mitotic phase – replicated DNA and cytoplasmic contents are separated; cell divides
Cytokinesis
Final stage of cell division
Cytoplasm divides to form two daughter cells
Interphase - Cell Cycle
G1 phase
Little visible change
Biochemically active
Accumulates building blocks for DNA, proteins, and energy for replication
S phase (Synthesis)
DNA is semi-condensed as chromatin
DNA replication produces sister chromatids attached at centromere
Centrosome is duplicated
G2 phase
Replenishes energy and synthesizes proteins for chromosome manipulation
Some organelles duplicated
Cytoskeleton dismantled to support mitotic spindle formation
Additional cell growth may occur
Mitotic Phase
Prophase
Chromosomes condense and become visible
Spindle fibers emerge from centrosomes
Nuclear envelope breaks down
Nucleolus disappears
Prometaphase
Chromosomes continue condensing
Kinetochores appear at centromeres
Spindle microtubules attach to kinetochores
Centrosomes move toward opposite poles
Metaphase
Mitotic spindle fully developed, centrosomes at opposite poles
Chromosomes aligned at metaphase plate
Each sister chromatid attached to spindle fiber from opposite pole
Anaphase
Cohesin proteins break down
Sister chromatids pulled toward opposite poles
Non-kinetochore spindle fibers elongate the cell
Telophase
Chromosomes arrive at poles and begin decondensing
Nuclear envelope reforms around each chromosome set
Mitotic spindle breaks down
Mitosis vs Meiosis
Mitosis
Single division
Produces two genetically identical diploid cells
For growth and repair
Meiosis
Two rounds of division
Produces four genetically unique haploid gametes
For sexual reproduction
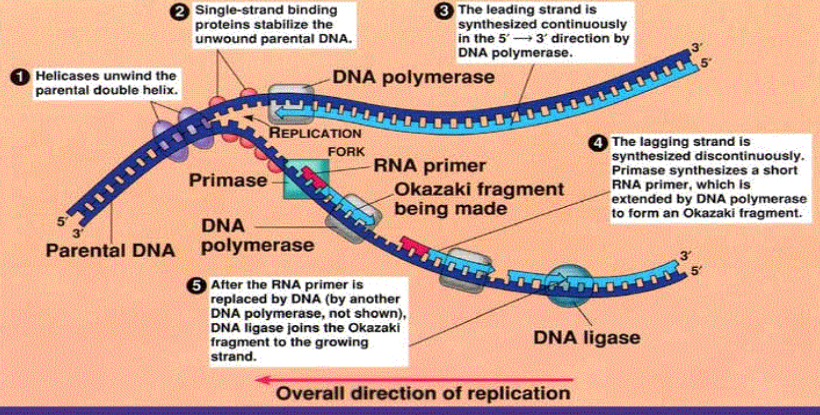
DNA Replication
Process producing two identical replicas from one original DNA molecule
Basis for biological inheritance
Semiconservative model
Parental DNA strands separate
Each strand serves as template for complementary daughter strand
Ensures daughter strands are identical to parent strand
Replication fork
Both strands replicate simultaneously
Leading strand – synthesized continuously 5’ → 3’
Lagging strand – synthesized discontinuously 5’ → 3’ as Okazaki fragments
Key enzymes and proteins
Helicase – unwinds parental DNA
SSBP (single-strand binding proteins) – stabilize single-stranded DNA
Primase – synthesizes RNA primers
DNA polymerase III – synthesizes new DNA 5’ → 3’
DNA polymerase I – replaces RNA primers with DNA
DNA ligase – joins Okazaki fragments
Replication direction
Always occurs 5’ → 3’
Leading strand continuous, lagging strand discontinuous
Summary of DNA Replication (photo)
Laws of Heredity and Genetic Variation (2)
Principle of Segregation
Characteristics of an organism are determined by alleles occurring in pairs
Allele pairs separate during gamete formation
Alleles randomly unite at fertilization
Example: in a 3-pair chromosome system, the two copies of each gene end up in different gametes
Principle of Independent Assortment
During meiosis, any allele of one gene may combine with any allele of another gene
Explains why offspring inherit combinations of traits that may differ from either parent
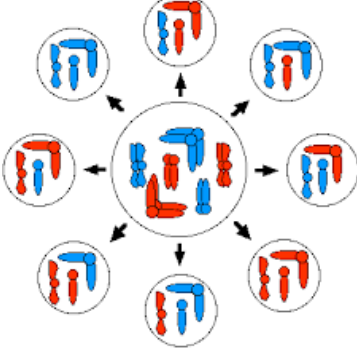
Genetic Variation
Differences in phenotype among individuals of the same species or population
Sources of genetic variation:
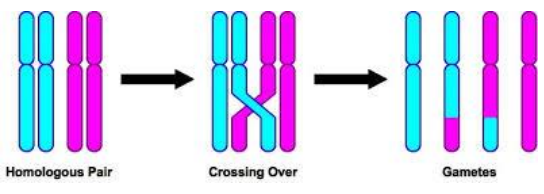
Crossing over during meiosis
Segregation and random fertilization
Independent assortment of alleles
Mutations
Crossing over - Genetic Variation (photo)
Segregation & random fertilisation - Genetic Variation (photo)
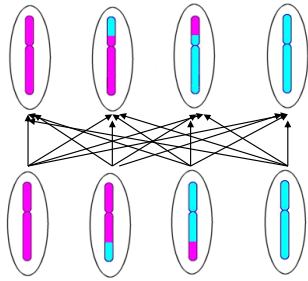
Independent assortment - Genetic Variation (photo)
Mutation - Genetic Variation (photo)
DNA Replication Errors
DNA polymerase duplicates DNA with high fidelity using strict base-pairing rules and proofreading
Replication errors occur about once per 10 million base pairs
DNA repair systems correct >99.9% of errors
DNA Repair Mechanisms
Direct Reversal Repair
Fixes DNA damage without excision
Examples:
UV-induced lesions repaired by photoreactivation
Alkylated bases repaired by enzymes like AGT and AlkB dioxygenases
Base Excision Repair (BER)
Removes and replaces damaged bases
Involves DNA glycosylases such as OGG1
Nucleotide Excision Repair (NER)
Repairs bulky lesions and cross-links from UV or chemicals
Removes damaged nucleotide fragments and synthesizes new DNA using the undamaged strand as template
Mismatch Repair (MMR)
Corrects base mismatches and insertion-deletion loops missed during replication
Steps:
Recognition of mismatch
Degradation of error-containing strand
Synthesis of correct DNA
Double-Strand Break Repair
Repairs DNA double-strand breaks (DSBs)
Two main pathways:
Homologous recombination (HR)
Non-homologous end joining (NHEJ)
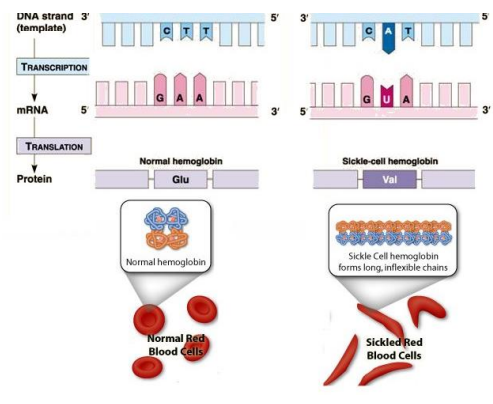
Genetic Code
Set of rules translating the four-letter DNA code into the 20 amino acids that make up proteins
Codons
Three-nucleotide sequences in DNA or RNA
Each codon corresponds to a specific amino acid or stop signal
64 possible codons: 61 code for amino acids, 3 are stop signals
Degeneracy of the code
Each codon specifies only one amino acid
Some amino acids are coded by more than one codon
Wobble Hypothesis
Base pairing rules are relaxed at the third codon position
A single tRNA can recognize more than one codon at this position
mtDNA vs Nuclear DNA Codon Usage
mtDNA
Stop codons: UAA, UAG, AGA, AGG
Tryptophan: UGA
Start codons: AUG, AUA, AUC, AUU
Nuclear DNA
Stop codons: UAA, UAG, UGA
Tryptophan: UGG
Arginine: AGA, AGG
Start codon: AUG
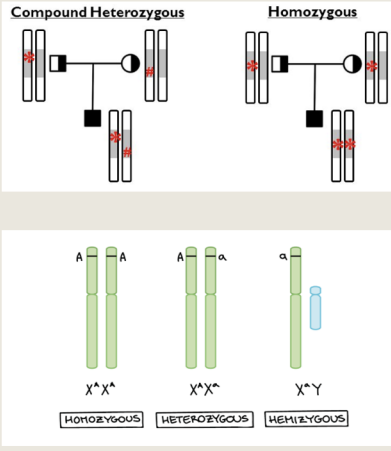
Single-Gene Disorders
Determined primarily by alleles at a single locus
Homozygous – pair of identical alleles at a locus
Heterozygous / Carrier – two different alleles at a locus
Compound heterozygote – two different mutant alleles of the same gene
Hemizygous – males with a single abnormal allele on the X chromosome, no second copy