DNA Biotechnology (2)
1/17
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
18 Terms
________________ is the preferred amplification technique in medicine for disease diagnosis and genetic analysis
PCR
- ex: bacterial infections, COVID tests, infectious diseases, viral infections
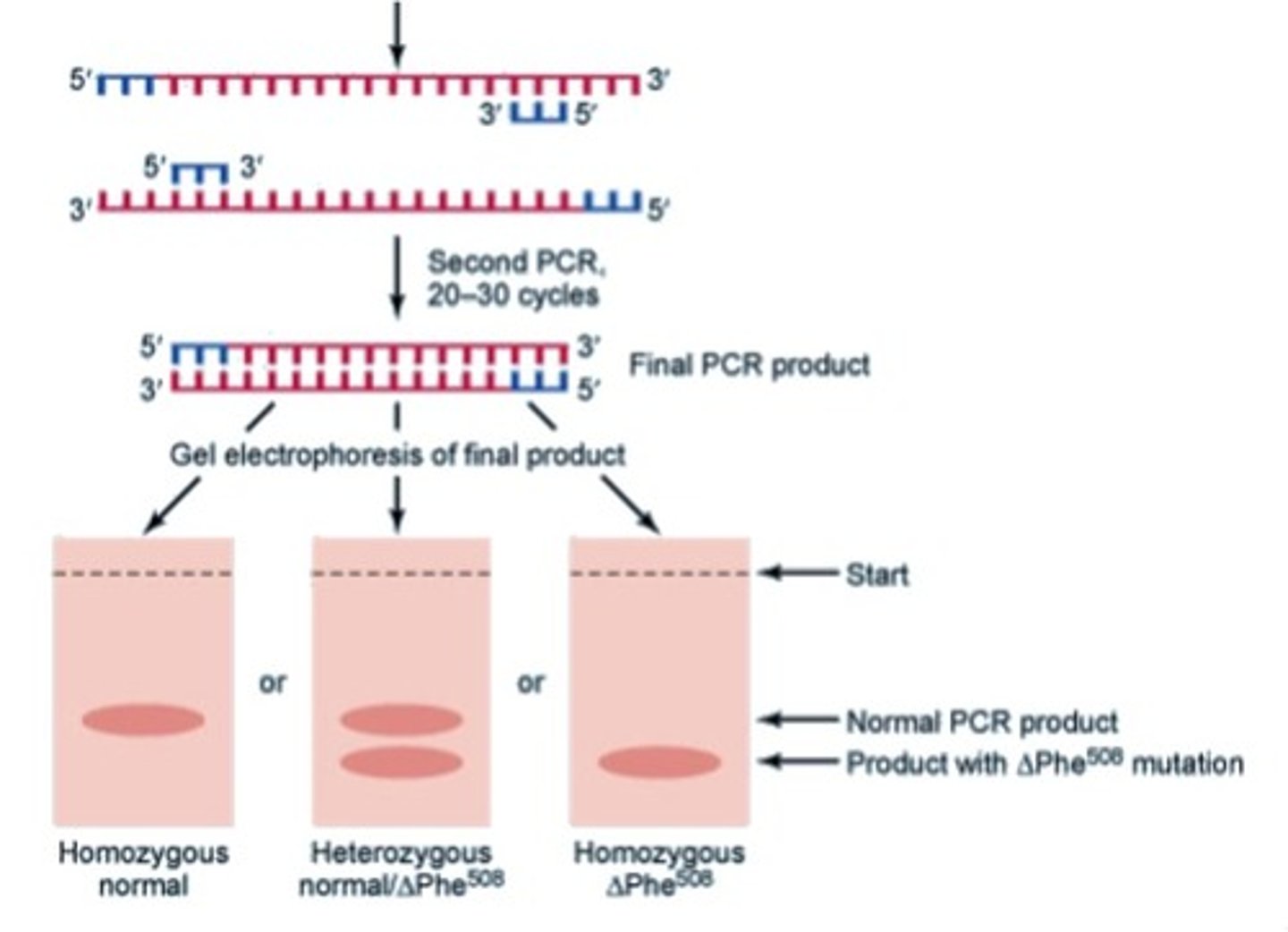
how can PCR be used to detect CF prenatally?
the mutant CFTR gene will be shorter than the normal version of the gene due to the loss of a phenylalanine residue
- using PCR to amplify the gene, then electrophoresis to visualize it, we can observe when the shorter version of the gene is present
- heterozygotes will show a normal AND mutant band, while mutant homozygotes will only have the shorter band
what are the 3 main blotting techniques, and what type of sample do each analyze?
- southern blot: DNA
- northern blot: RNA
- western blot: protein
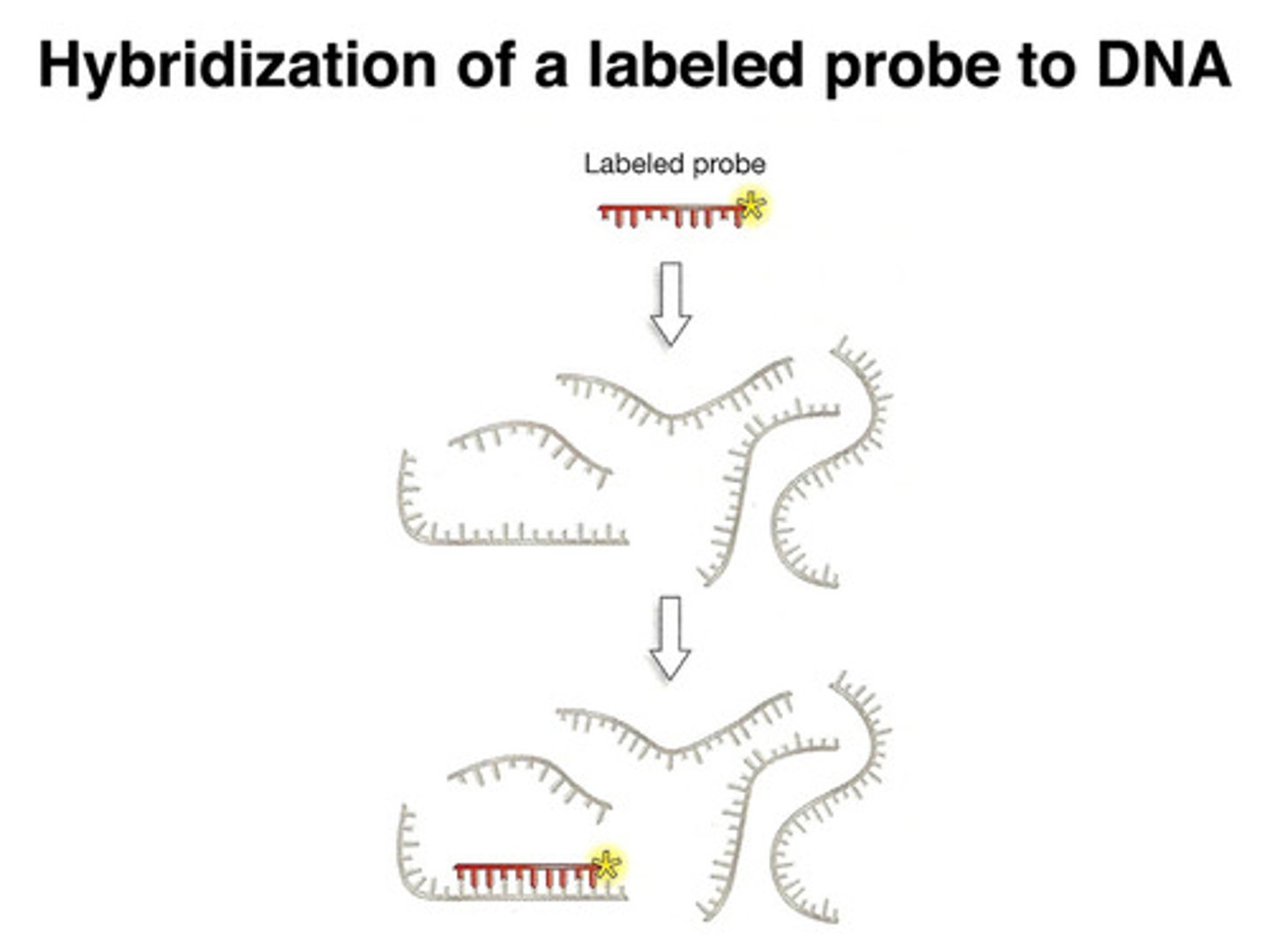
what are probes? how does DNA hybridization occur? what is this process used for?
a short piece of ssDNA or RNA labeled with a radioisotope, like 32P
- the sequence of a probe is complementary to a sequence in the target DNA
- probes are used in screening to identify which band on a gel contains the target DNA
how does northern blotting work? what is the application of technique?
① mRNAs in a sample are separated by electrophoresis
② the mRNAs are then transferred to a membrane and hybridized with a radiolabeled probe
③ the bands obtained by autoradiography give a measure of the amount and size of the mRNAs in the sample
application: identifying specific RNA sequences and measuring their abundance in a sample
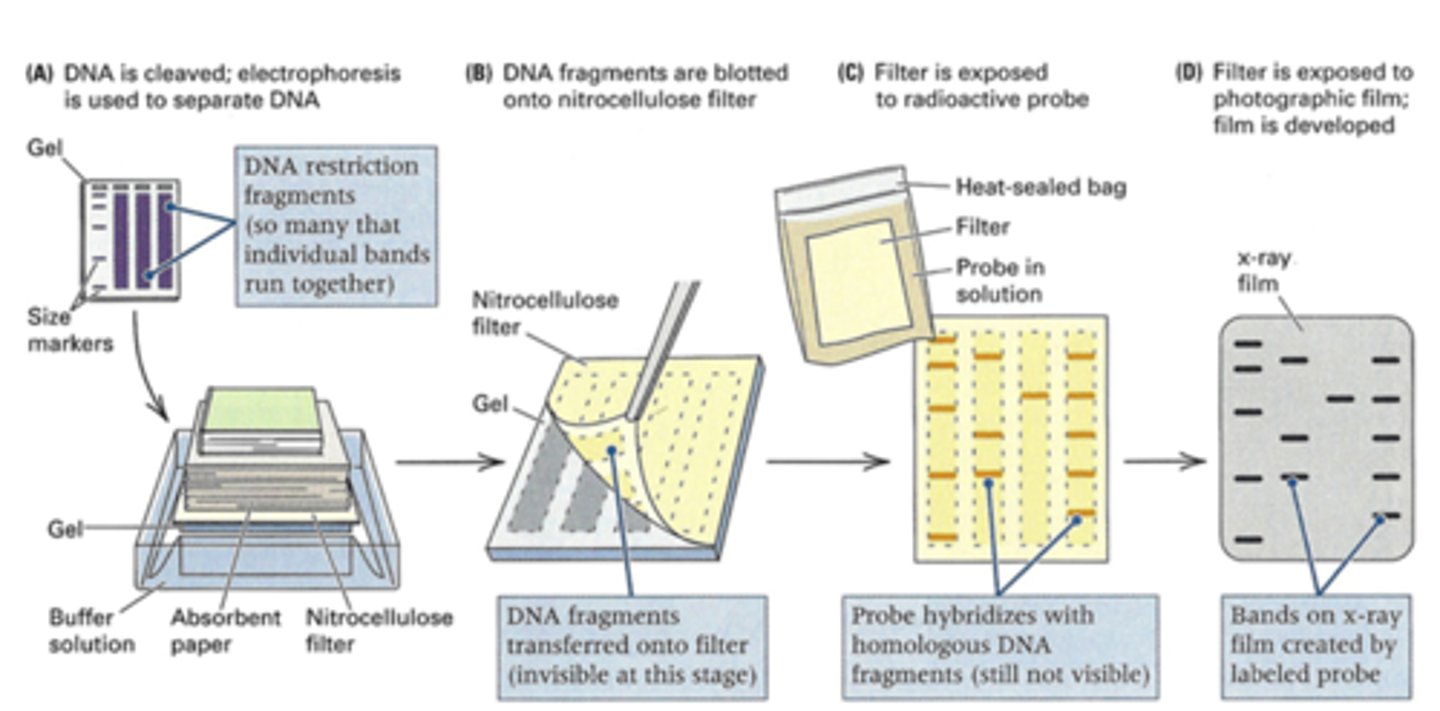
how does southern blotting work?
combines the use of restriction enzymes, electrophoresis, and DNA probes to generate, separate, and detect pieces of DNA
① DNA is extracted from a patient's WBCs
② DNA is cleaved into fragments using restriction enzymes
③ the fragments are separated by size using agarose DNA gel electrophoresis, then blotted onto a nitrocellulose membrane for analysis
④ hybridize the membrane using a radioactive probe to detect the fragments
what are the applications of using PCR for DNA fingerprinting?
- compare DNA from 2 different individuals
- compare DNA of 2 different species
- compare samples of blood, hair, or tissue found at a crime scene
T/F: the human genome is mostly the same in all individuals
TRUE
- genetic variations account for about 0.01% of each person's DNA
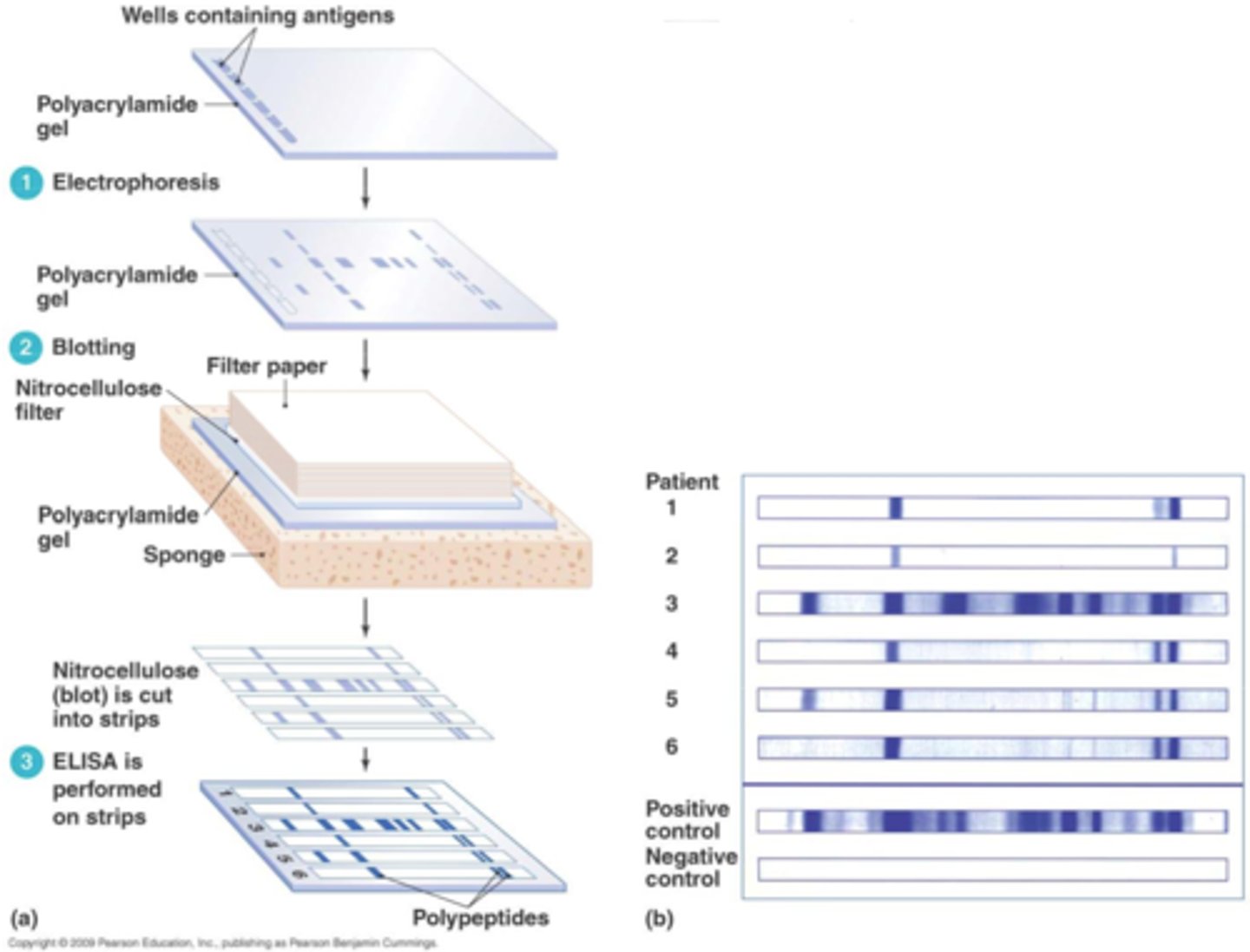
how do western blots work?
① proteins are separated by electrophoresis
② blotted onto a membrane
③ the protein-blotted membrane is hybridized with an Ab
④ hybridization produces a band on the membrane at the location of its Ag
T/F: the amount of protein in cells always directly correlates to the amount of mRNA present
FALSE
- NOT always correlated!
______________ are used to detect and quantify specific proteins and to determine posttranslational modifications
labeled Abs
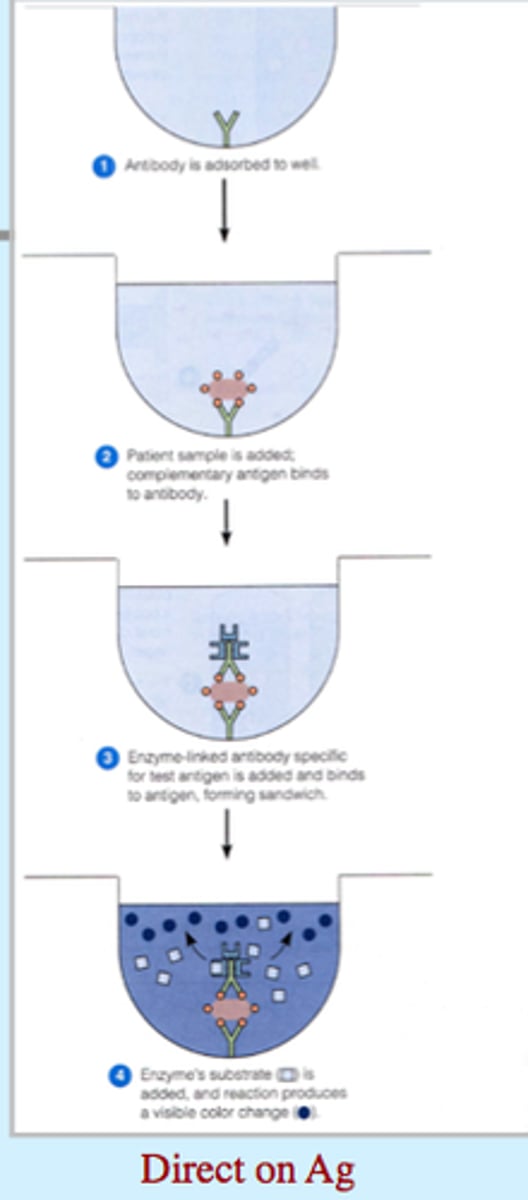
how do enzyme-lined immunosorbent assays (ELISA) work?
- the Ag (protein) is bound to the plastic of the dish
- the probe, which is an Ab that's specific to the protein being measured, is covalently bound to an enzyme
- the enzyme produces a colored product when it is exposed to its substrate
- the amount of color produced is proportional to the amount of Ab present and, indirectly, to the amount of protein in a sample
what types of biological tests are used to detect HIV?
- ELISAs are used as a primary screening tool
- western blots are used as a more specific, confirmatory test
- PCR-based testing is more useful in the first few months after exposure, as it directly detects the viral nucleic acids
how are DNA mutations detected?
- direct sequencing
- DNA hybridization
- restriction enzyme digestion methods → if a mutation alters the restriction site for a specific enzyme, the size of the PCR product will change (seen with electrophoresis)!
what type of probe is used to detect the presence of the sickle cell mutation in the β-globin gene?
a synthetic, radiolabeled allele-specific oligonucleotide (ASO)
- complementary to the point mutation (GAG → GTG) at codon 6 in patients with the β(s) gene
- DNA is taken from the patient's WBCs, amplified, denatured, then attached to a membrane before being exposed to the probe
in sickle cell anemia, the sequence alteration caused by the point mutation disrupts the recognition site of what restriction enzyme? what type of diagnostic tool is this?
endonuclease MstII
- site sequence: CCTNAGG
- normal DNA digested with this enzyme yields a 1.15-kb fragment, while the mutated DNA yields a 1.35-kb fragment
- this is an example of a restriction fragment length polymorphism (RFLP) diagnostic tool
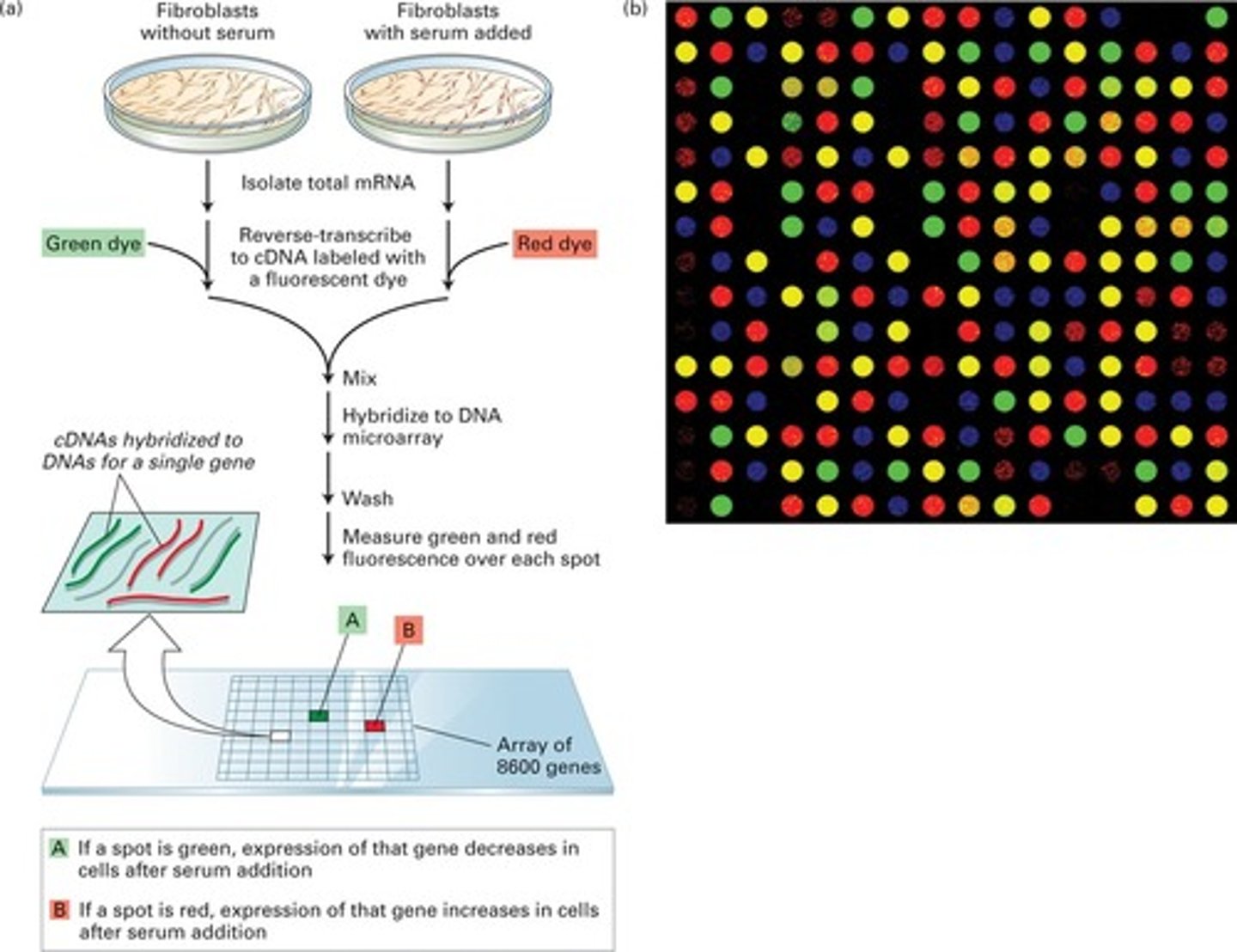
what are DNA microarrays used for? how do they work?
to determine the differing patterns of gene expression in 2 different cell types (ex: normal vs. cancerous); important for personalized medicine!
- analysis of mRNA and protein products of gene expression
- population of mRNA molecules from a particular cell type is converted to cDNA and labeled with a fluorescent tag
- this mixture is then exposed to a glass slide (chip) containing thousands of different genes
what are some clinical applications of microarray-based diagnostic tests?
genotyping genes of drug metabolizing enzymes
- microarray-based genotyping → amplichip CYP450 test
- allelic variants of both CYP2D6 and CYP2C19!