Unit 6 Molecular Biology
1/68
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
69 Terms
Fridrich Meischer
First to discover DNA.
Extracted white blood cells from blood and lysed the cells.
Called it ‘Nuclein’. (later to be called nucleic acids)
Phoebus Levene
Determined nucleotide structure: Base, Sugar, Phosphate Group
Proposed the tetranucleotide hypothesis, which claimed DNA was a simple, repeating sequence and therefore too uniform to carry genetic information. That proved to be incorrect)
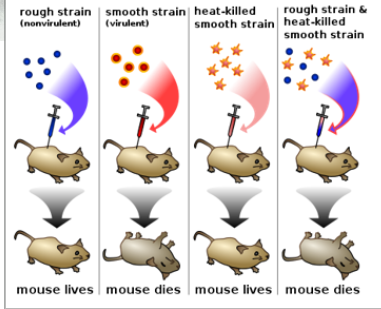
Griffith Experiment
Griffith was working with bacteria that caused pneumonia.
S (Smooth) strain of bacteria would kill the mouse.
R (Rough) strain would not kill the mouse.
Boiled S strain Injected boiled S strain into the mouse and the mouse was ok!
Mixed boiled S strain and R strain and injected it into the mouse. Killed the mouse.
-> He found living S strain in the dead mice
Came up with Transformation Principle.
Transformation Principle
Griffith Experiment
Bacteria can transfer genetic material through transformation.
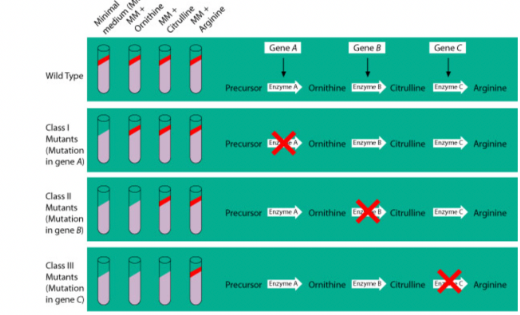
Beadle and Tatum
Showed that genes control the production of enzymes.
Used bread mold (Neurospora crassa).
X-rays were used to create mutations.
Mutant molds could no longer make certain molecules needed to grow.
When a missing nutrient was added back, growth resumed.
Conclusion:
Each gene is responsible for making one enzyme (later refined to one gene → one polypeptide).
One gene-one enzyme hypothesis
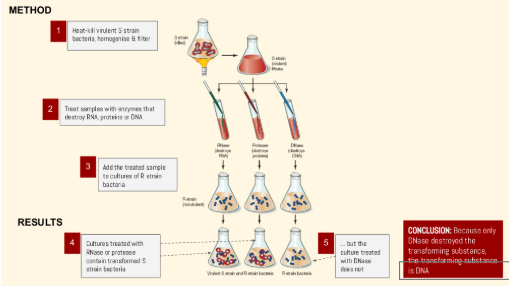
Avery, McLeod, McCarty
Built on the work of Griffith to show that DNA (not protein) was the transforming principle
THE PROCESS:
Isolated DNA, RNA, and proteins from heat-killed virulent S. pneumoniae bacteria and tested which ones could transform non-virulent R-strain bacteria.
Only DNA was able to transform the non-virulent bacteria into the virulent form.
Degradation Tests: When DNA was treated with DNA-digesting enzymes (DNase), transformation did not occur, but treatments with protease (breaking down proteins) or RNase (breaking down RNA) transformation occured.
Central Dogma
DNA is transcribed into RNA and translated into protein
DNA —> RNA —> Protein
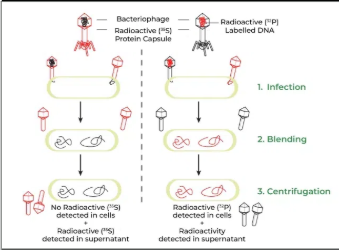
Hershey and Chase
Waring Blender Experiment
Two groups of bacteriophages were labeled:
35S (sulfur) labeled the protein coat because sulfur is in proteins, not DNA.
32P (phosphorus) labeled the DNA because phosphorus is in DNA, not proteins.
The bacteriophages infected bacteria.
A blender separated the viral coats from the bacteria.
Centrifugation separated the parts by density.
The bacteria contained 32P, showing that DNA entered the cells, not protein.
James Watson and Francis Crick
Proposed the double-helix structure of DNA in 1953
Showed that DNA consists of two antiparallel strands with complementary base pairing (A–T and C–G)
Heavily relied on Rosalind Franklin
Maurice Wilkins
Who “shared” Franklin’s photo with James Watson and Francis Crick.
Watson and Crick created the 1st accurate model of DNA with Franklin’s work.
Rosalind Franklin
Scientists knew DNA was made of nucleotides, but didn’t understand total structure.
Studied x-ray diffraction and was studying DNA.
Took “Photo 51”
Linus Pauling
Alpha-Helix Structure: In the 1950s, Pauling proposed the alpha-helix as a key structural feature of proteins, a breakthrough that helped understand protein folding. He also was the part of the discovery of β sheets.
Protein Structure: He emphasized the role of hydrogen bonds in protein stability w/in secondary structure.
DNA Structure Hypothesis: Pauling attempted to model the structure of DNA, proposing a triple-helix model in 1953, which was later shown to be incorrect.
DNA Structure
The monomers of DNA are nucleotides.
Nucleotides are made of base+sugar+phosphate. Phosphate Group, PO43-, deoxyribose sugar, Nitrogenous base: purine or pyrimidine
Eukaryotic Chromosome
Shape - Linear
Size - Large
Number - Multiple
Location - Nucleus
Storage proteins - Histones
Prokaryotic Chromosome
Shape - Circular
Size - Small
Number - Single
Location - Nucleoid (region in cytoplasm)
Storage proteins - Nucleoid associated proteins (supercoiling)
Double Bonds
A and T
Triple Bonds
G and C
DNA Bases
A, G, T, C
RNA Bases
A, G, U, C
Pyrimidines
Single Rings
Cytosine
Thymine (DNA only)
Uracil (RNA only)
Purines
Double Rings
Guanine
Adenine
Bases Bonding
Sugar-phosphate backbone
Bases bound with hydrogen bonds
Pyrimidines bound to purines
Chargaff’s Rule
% A = % T
% G = % C
He figured out base pairing
This was crucial information that helped Watson & Crick
This is true for all species
Double Helix
Two chains of nucleotides in a twisted ladder structure called a
Phosphodiester Bonds (covalent)
Backbone is composed of repeating deoxyribose sugar and phosphate bonded connected by
Hydrogen Bonds
Nitrogenous bases form the rungs of the ladder and are connected by
Antiparallel
DNA runs 5’ to 3’ and 3’ to 5’.
One end is 5’ and the other 3’.
5’ end is phosphate
→ 5’ PO43-
3’ end is sugar
→ 3’OH–
Major and Minor Grooves
The two unequal spaces that run along the outside of the DNA double helix.
Major Groove - Larger
Minor Groove - Smaller
Chromosome
Tightly packed DNA and protein structure. DNA takes on this form as the cell prepares to undergo division. (“butterfly”)
Histone
Protein molecule that DNA wraps itself around. (Spools)
Positively charged because they contain a lot of arginine and lysine, both of which carry a net positive charge
Then bind to negatively charged phosphate groups in the sugar phosphate backbone of DNA
Chromatin
Thin thread of DNA. Consists of DNA and histones. It will condense before mitosis. (“spaghetti”)
Phosphate groups (charged)
Negatively charged
8 Histones
DNA coils around
Nucleosomes
DNA is coiled around 8 histone proteins to form
Chromatin (form)
Nucleosomes are coiled again to form
DNA Packaging Steps
DNA wraps around histone proteins forming beads on string” called nucleosomes.
Nucleosomes further coil and condense/gather to form chromatin.
Chromatin fibers can unwind for DNA replication and transcription.
Plasmids
Small, circular pieces of double-stranded DNA found in prokaryotes and some eukaryotes.
Often carry helpful genes like antibiotic resistance.
Used as vectors to carry new genes into cells for gene cloning, genetic engineering (such as making insulin), and gene therapy.
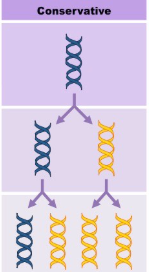
Conservation (Replication)
The parent double-helix DNA is copied in its entirety, and the new cell’s DNA is entirely a copy of the old.
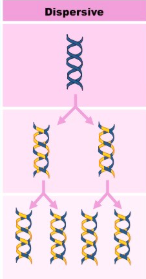
Dispersive (Replication)
DNA is chopped up into pieces; these little pieces are copied and then reassembled in combination with the old pieces.
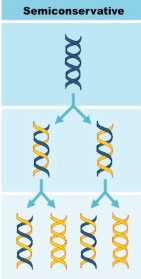
Semiconservative (Replication)
DNA Replication is
The double-stranded DNA separate from their helix shape, and each makes a copy of itself. The new cells then contain one strand from the parent cell and one newly synthesized strand.
Meselson & Stahl Experiment
“The Most Beautiful Experiment in Molecular Biology”
How did we know the DNA Replication is semiconservative
Meselson & Stahl
Bacteria were first grown in ¹⁵N (heavy nitrogen) so their DNA became heavy.
A sample of this ¹⁵N DNA was saved.
The rest were moved to ¹⁴N (light nitrogen) so new DNA would be light.
Samples were taken after each time the bacteria doubled.
A sample grown only in ¹⁴N was used for comparison.
DNA was taken out and mixed with a strong salt solution.
The samples were spun fast in a centrifuge.
Heavy DNA (¹⁵N) sank lower than light DNA (¹⁴N), showing differences in DNA.
S Phase
DNA replication occurs in
Initiation
Origin of replication: Specific DNA sequences are recognized by initiator proteins.
Helicase unwinds the DNA: Unzips the double helix, creating a replication bubble with two replication forks.
Primer Synthesis
Primase synthesizes short RNA primers on the single-stranded DNA templates to provide a starting point for DNA synthesis.
Elongation
DNA polymerase III adds nucleotides to the 3' end of the primer, synthesizing the new DNA strand in the 5' to 3' direction.
On the leading strand, synthesis is continuous. On the lagging strand, DNA is synthesized in Okazaki fragments.
Primer Removal
DNA polymerase I removes RNA primers and fills in the gaps with DNA.
Ligation
DNA ligase seals the nicks between adjacent DNA fragments, joining the newly synthesized DNA strands into a continuous strand.
Origin of Replication
Location where replication begins. This is where the unzipping begins.
Euk: multiple (b/c large genome)
Prok: single
Replication Fork
Y-shaped area where DNA is being copied. The enzyme helicase unwinds the double-stranded DNA, creating the fork so new DNA strands can be made.
Helicase
Unwinds DNA by breaking hydrogen bonds between bases
Requires ATP (ATP hydrolysis)
Topisomerase
Relieves torsional strain/supercoiling caused by unwinding
Single stranded binding proteins
Binds to ssDNA to prevent it from reannealing (coming back to double strands)
Stabilize strands to keep them single-stranded, no enzymatic activity
Primase
Enzyme that moves 5’ to the 3’
Lays down RNA primers by reading ssDNA
Creates short RNA sequence (about 10 to 15 nucleotides long)
Primers
Necessary because DNA Pol III needs a free 3'OH to add nucleotides to the growing chain.
DNA Polymerase 3
Enzyme that binds RNA primers and adds nucleotides to elongate the DNA strand in the 5’ to 3’ direction.
It forms a phosphodiester bond by joining the 3' OH of the growing DNA strand to the 5' phosphate of the new nucleotide.
VERY ACCURATE
DNA Polymerase 1
Replaces RNA primers with DNA and proofreading
Ligase
Seals nicks between DNA fragments
Forms phosphodiester bonds between the sugar-phosphate backbones of adjacent nucleotides. Requires ATP.
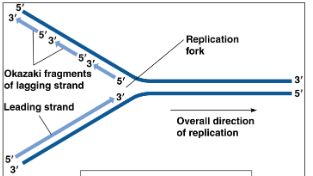
Leading strand
Made continuously, DNA Pol III moves towards replication fork.
5’ to 3’ direction
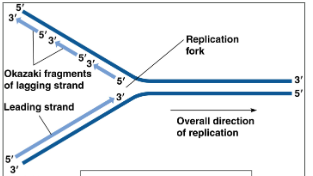
Lagging strand
Made discontinuously, DNA Pol III moves away from replication fork. Creates small sections known as Okazaki fragments
3’ to 5’ direction
3’ to 5’
DNA polymerase can only read and add nucelotides to the template of
5’ to 3’
The new strand grows
5' to 3' Exonuclease Activity (DNA Poly 1)
Removes RNA primers. removes RNA primers by cleaving the RNA bases in the 5' to 3' direction
3' to 5' Exonuclease Activity (DNA Poly 1)
Proofreading function, moves backwards along the newly synthesized DNA (in the 3' to 5' direction)
Removes nucleotides one at a time from the end of a DNA strand, fixing mistakes during DNA replication
Endonuclease activity
Cuts nucleotides in the middle of a strand rather than at the ends, can cut out damaged or mismatched DNA
Methylation (CH3)
DNA is after it’s replicated.
Happens to control gene activity. It can turn genes off or down, helping the cell know which genes to use and which not to use.
- Prok: usually A is methylated
- Euk: usually C is methylated
Excision Repair
Fixes damaged DNA, like damage from UV light or chemicals (thymine dimers). The damaged section is cut out and replaced with correct DNA.
Mismatch Repair
Happens after DNA replication.
The cell finds and fixes mistakes that were missed by proofreading by cutting out a section of the new strand and replacing it.
Xeroderma pigmentosum
Rare Genetic Disorder
Defective Excision Repair because of a mutation in one of the genes responsible for excision repair.
Recall: UV radiation from sun can cause Thymine-Thymine dimers.
Can’t go out in sunlight
Increased skin cancers/cataracts
1 in 1,000,000 million