Lecture 18 - Micro RNA and siRNA reg of gene expression
1/28
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
29 Terms
What are the two main classes of small RNAs involved in RNA interference?
Small interfering RNAs (siRNAs)
MicroRNAs (miRNAs). Both are short (~20–30 nt)
Not ‘junk DNA’ non-coding RNAs that regulate gene expression by promoting mRNA degradation or inhibiting translation.
What is the biological role of RNA interference in plants and animals?
In plants/insects: protects against RNA viruses.
In animals: suppresses transposable elements that replicate via RNA intermediates.
This represents an evolutionarily conserved antiviral and genome-defense mechanism that uses RNA degradation to destroy foreign RNA.
What are transposons?
Transposons, or jumping genes, are segments of DNA that can move within the genome, potentially causing mutations or altering the cell's genome structure.
They often replicate through RNA intermediates, affecting gene expression and genome stability.
What is the general mechanism of how transposons work?
Operate by copying their DNA to RNA, which is reverse-transcribed back to DNA and inserted at another site in the genome
dsRNA can be targeted by RNA interference
Protects the genome against transposons
What are the two methods of transposition?
Cut-and-paste mechanism: involves the transposon being excised from one location and inserted into another
Copy-and-paste mechanism: creates an additional copy that inserts into a new location without removing the original.

Who discovered RNA interference and when?
Andrew Fire and Craig Mello in C. elegans (Nature, 1998).
Found that double-stranded RNA (dsRNA) silenced expression of a gene for muscle function more effectively than antisense RNA, causing it to twitch in a similar way to worms that lacked a functional muscle protein gene.
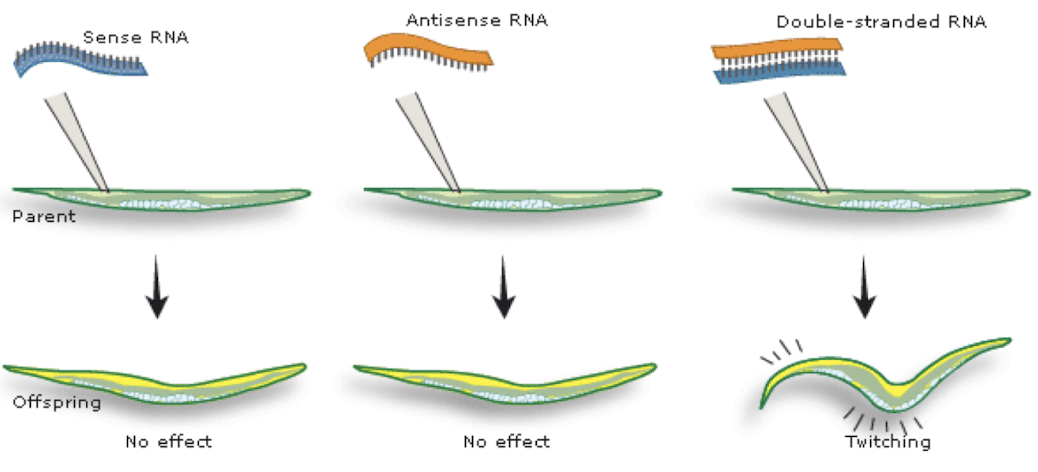
Why was dsRNA more effective than antisense RNA in gene silencing experiments?
dsRNA triggers a cellular defence mechanism involving Dicer and the RISC complex, leading to sequence-specific mRNA degradation, while antisense RNA only blocks translation passively
What was the first discovered miRNA, and in what organism?
The first miRNA, lin-4, was discovered in C. elegans
It did not encode a protein but produced a small RNA that imperfectly base-paired to complementary sequences on target mRNAs

What is let-7 and why was its discovery significant?
let-7 was discovered in C. elegans (2000) and found to be conserved from flies to humans.
It established miRNAs as a universal mechanism of post-transcriptional regulation across species.
How do miRNAs contribute to developmental robustness?
miRNAs fine-tune gene expression by buffering transcriptional noise, ensuring stable developmental outcomes even under environmental fluctuations — a concept known as canalisation.
Approximately how many miRNAs exist in humans, and what fraction of genes do they regulate?
There are >2000 known human miRNAs, regulating up to 1/3 of all mRNAs — over 1% of the human genome is dedicated to miRNA sequences.
Are highly conserved
List 5 major cellular processes regulated by miRNAs.
Developmental timing - i.e. control of genes during early development
Tissue growth
Tumour suppression
Cell differentiation
Apoptosis.
functionally similar to siRNA but unique in means of biogenesis
From where are functional miRNAs transcribed in the genome?
From introns of protein-coding genes,
From introns/exons of non-coding genes, or
From intergenic regions under independent promoters (control themselves, all are transcribed mainly by RNA polymerase II.
What are some key points about the transcription of miRNA in eukaryotes?
miRNAs originate from monocistronic, dicistronic or polycistronic primary transcripts (pri-miRNAs) that are generated by RNA Pol II in all eukaryotes
Human pre-miRNA’s are capped and polyadenylated – unique properties of RNA pol II transcripts
Many miRNAs are regulated by important TFs such as cNF-kB, c-Myc and p53
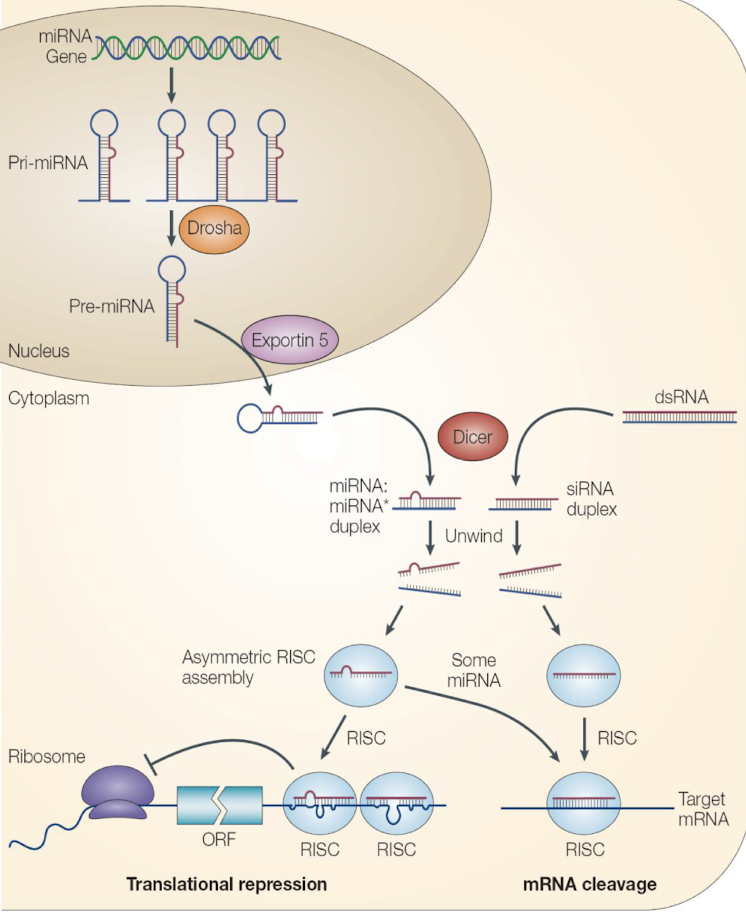
What is the role of Drosha and DGCR8 in miRNA biogenesis?
In the nucleus, Drosha is an RNase III enzyme and its cofactor DGCR8 “crop” the primary miRNA transcript (pri-miRNA) into a precursor miRNA (pre-miRNA) with a stem-loop structure.

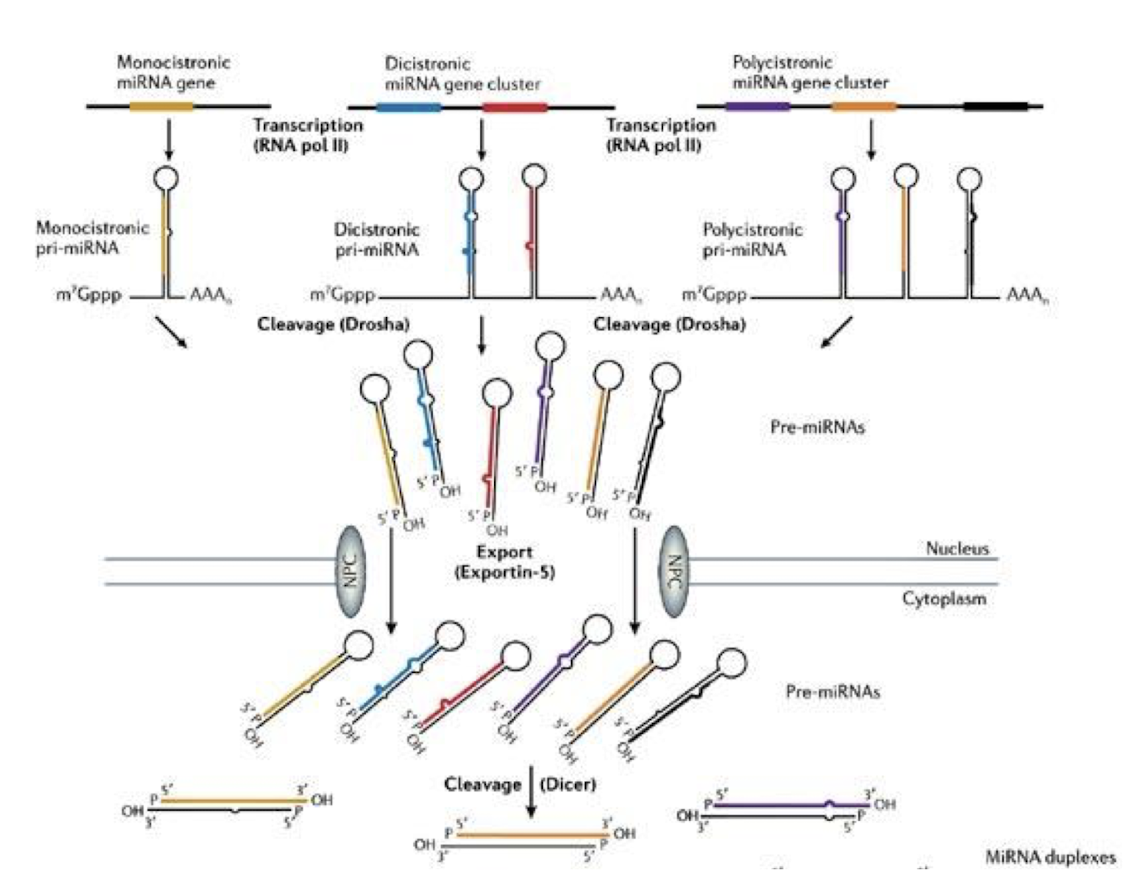
How is pre-miRNA exported to the cytoplasm?
Via Exportin-5 and Ran-GTP, which recognise the pre-miRNA’s 2-nt 3′ overhang and facilitate nuclear export.
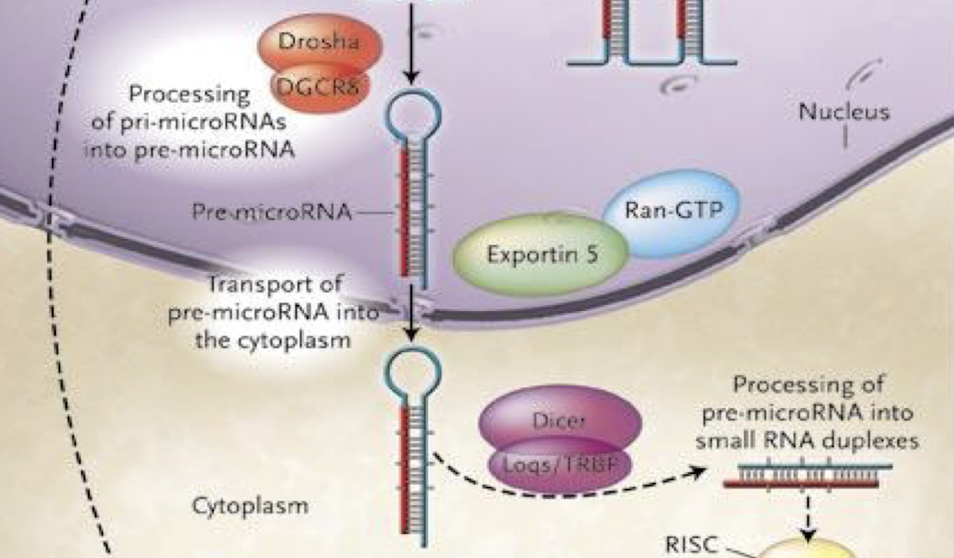
What is Ran-GTP important for?
Ran-GTP is crucial for the transport of RNA and proteins across the nuclear membrane, specifically for facilitating the export of pre-miRNA and other cargoes from the nucleus to the cytoplasm.
Describe Dicer’s role in miRNA maturation.
In the cytoplasm, Dicer (RNAse III endonuclease enzyme) processes pre-miRNA into a ~22-nt RNA duplex with 3′ 2-nt overhangs, cleaves the stem loop
One strand (the guide strand) is incorporated into RISC, while the other (the passenger strand) is degraded.
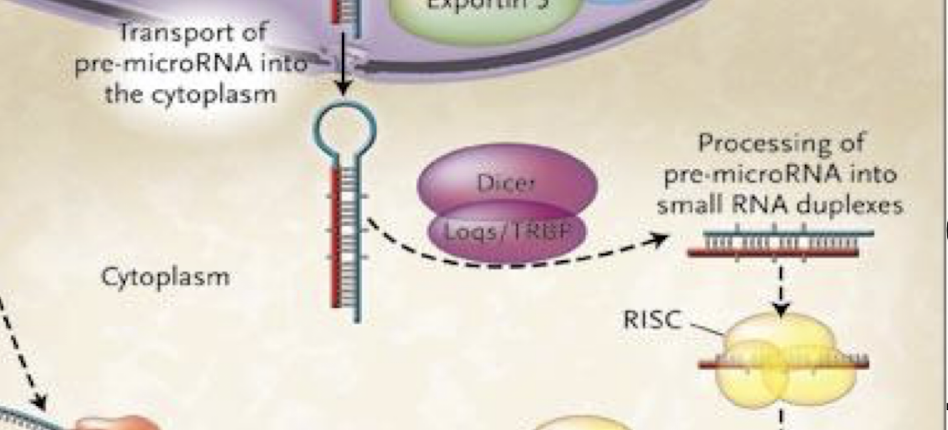
What is the role of the Argonaute (AGO) protein?
AGO binds to the guide strand and forms the RNA-induced silencing complex (RISC) by inducing strand dissociation and degradation
It mediates either mRNA cleavage (if base-pairing is perfect) or translation repression (if imperfect).

Where do miRNAs typically bind on the target mRNA?
The 3′ untranslated region (3′ UTR), though binding in the 5′ UTR or coding region can occur in some cases
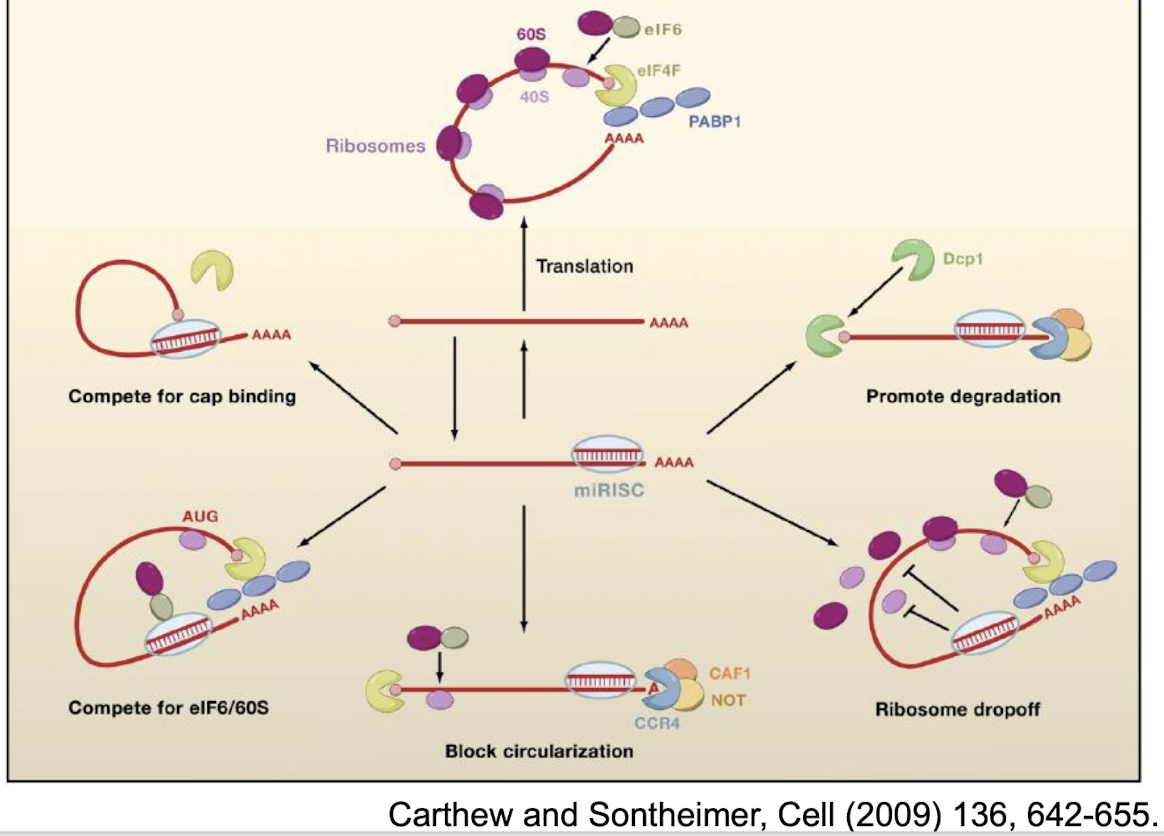
What are some examples of the inhibitory mechanisms by miRNA on translation?
Compete for cap binding i.e. acylation factors by recruiting deadenylases and decapping enzymes
Compete for eIF6/ 60S
Block circularisation
Ribosome drop-off
Promote degradation
What are the shared components between siRNA and miRNA pathways?
Both involve:
A double-stranded RNA trigger,
Dicer for RNA processing, and
Argonaute-containing RISC complex for effector function i.e. transcriptional repression / mRNA cleavage
How are miRNAs implicated in cancer?
Many miRNAs are located in cancer-associated genomic regions and are deregulated in tumours.
Involved in a variety of haematological malignancies like leukemia (AML,CML and CLL) as well as diffuse large B-cell lymphoma.
Some act as oncogenes (oncomiRs) or tumour suppressors, influencing proliferation, apoptosis, and metastasis.
Give examples of oncogenic and tumour-suppressor miRNAs:
Oncogenic miRNAs: e.g. miR-155, miR-17–92 cluster → promote tumour growth.
Tumour-suppressor miRNAs: miR-15a, miR-16, miR-34, let-7 → inhibit tumourigenesis.
What are two therapeutic strategies involving miRNAs?
miRNA antagonists (antagomiRs): inhibit overactive oncogenic miRNAs
miRNA replacement therapy: restore expression of lost tumour-suppressor miRNAs
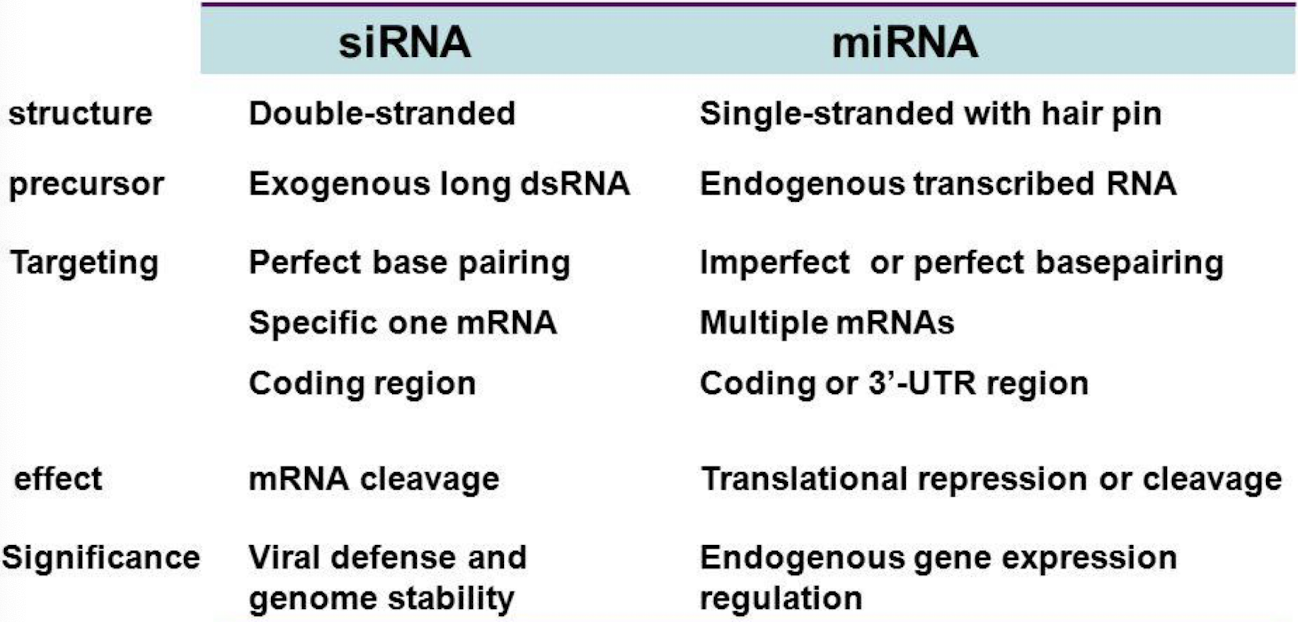
What is the difference between siRNA and miRNA origin?
siRNA: derived from exogenous dsRNA (viruses, transposons, synthetic molecules).
miRNA: encoded endogenously by the genome and transcribed by RNA Pol II.
Why are Dicer and Argonaute considered part of the RNA-induced silencing machinery (RISC)?
They process and direct the guide strand to complementary mRNA targets, forming the core catalytic components of RISC, which mediates post-transcriptional gene silencing.
How can synthetic siRNAs be used experimentally?
Researchers design siRNAs complementary to target mRNAs to knock down specific genes, providing a reversible and sequence-specific alternative to genetic knockouts.
How can we summarise the differences between siRNA and miRNA?