innate immunity
1/21
Earn XP
Description and Tags
week 1 ctb
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
22 Terms
immune system
provides protection against infectious disease, but also relevant to neoplasia, allergy, autoimmune disease, transplantation, diagnostics, etc
spread throughout body so a functional (cellular) approach is more meaningful than an anatomical one
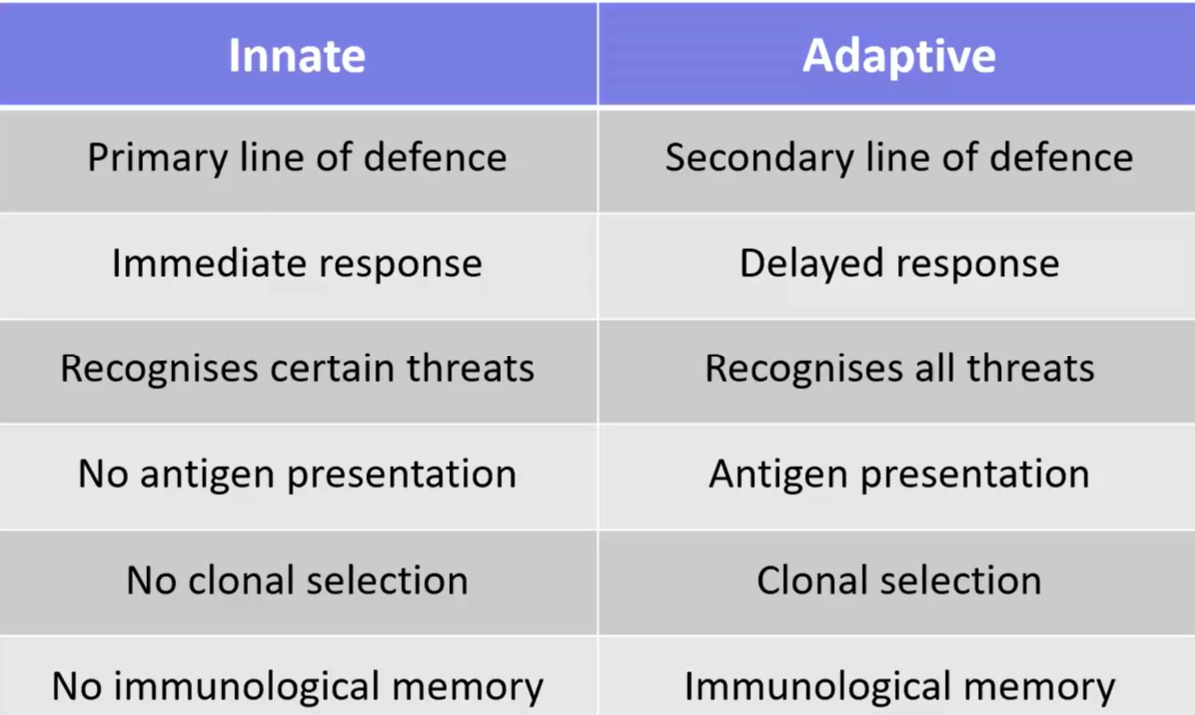
immune function can be divided into innate and adaptive immunity although these are interlinked
antigen= any molecule capable of inducing immune response but is often thought in terms of specific adaptive immune response
innate vs adaptive table
innate immunity
defences against disease that are naturally present rather than induced by prior exposure to a pathogen
non-specific defence mechanisms that become active immediately (within hours) upon exposure
not long-lasting and without immunological memory
relatively poor amplification or regulation of response leading to threat to self-antigens
evolutionarily ancient and therefore widespread among species: pathogens have immunity too
components of the innate immune system
physical barriers: epithelia and secretions
leukocytes
phagocytic cells
granulocytes
NK cells
plasma proteins: humoral response
integration of innate and adaptive immune responses
innate and adaptive immune responses are not isolated processes but act together to optimise response to pathogens
adaptive immune response is very powerful/specific but many pathogens would kill in the days needed for a good adaptive response in the absence of non-specific innate immune responses
barrier immunity
physical barriers
skin
mucus
respiratory cilia
biochemical barriers
sebaceous secretions in skin
lysozyme in tears
gastric acidity
commensal organisms
most infectious agents enter via the mucosal surfaces of
nasopharynx
respiratory tract
gastro-intestinal tract
genito-urinary tract
epithelial defences
interior epithelial surfaces are covered with mucus containing mucins (help prevent pathogens from adhering and facilitate their clearance by cilia)
peptides in mucus called defensins kill/inhibit growth of pathogens
cells of the blood
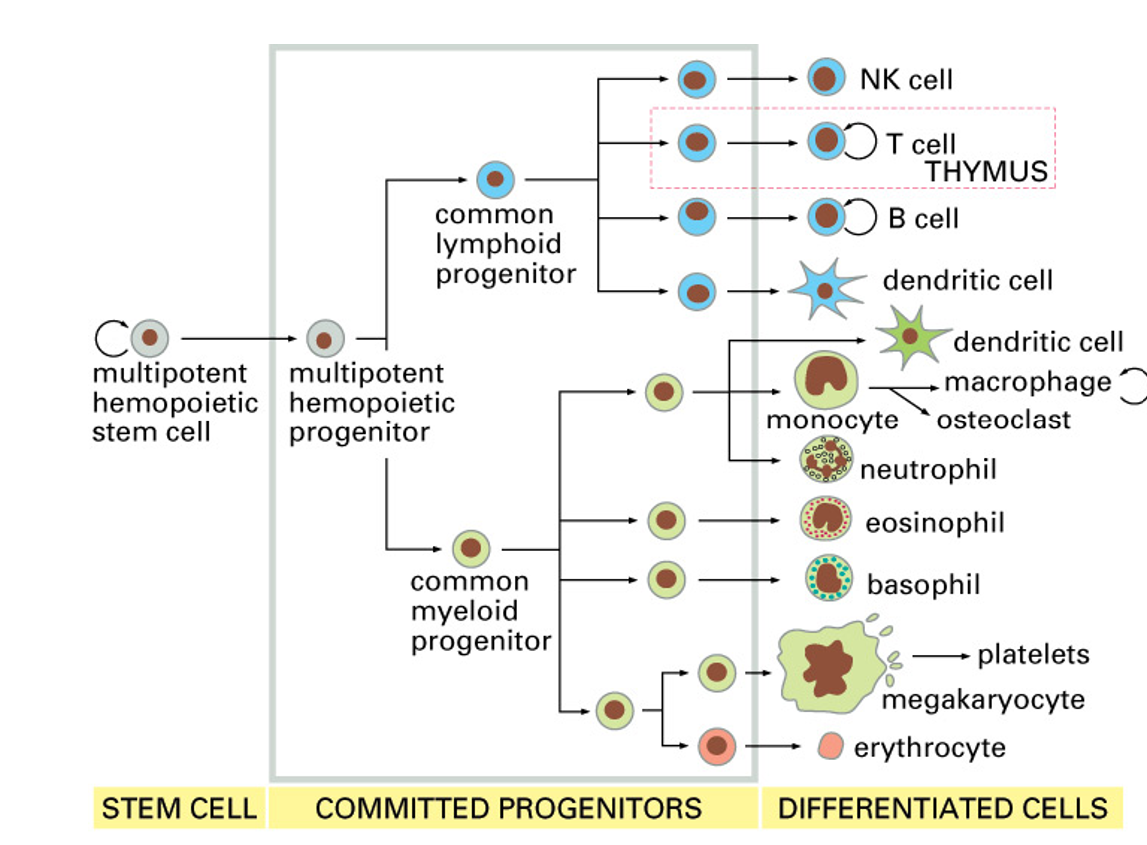
myeloid lineage cells (main cells involved in innate immunity)
phagocytosis (including an oxidative burst)
chemotaxis/adherence of microbe to phagocyte
ingestion of microbe by phagocyte
formation of a phagosome
fusion of phagosome with lysosome to form phagolysosome
digestion of ingested microbe by enzymes
formation of residual body containing indigestible material
discharge of waste materials
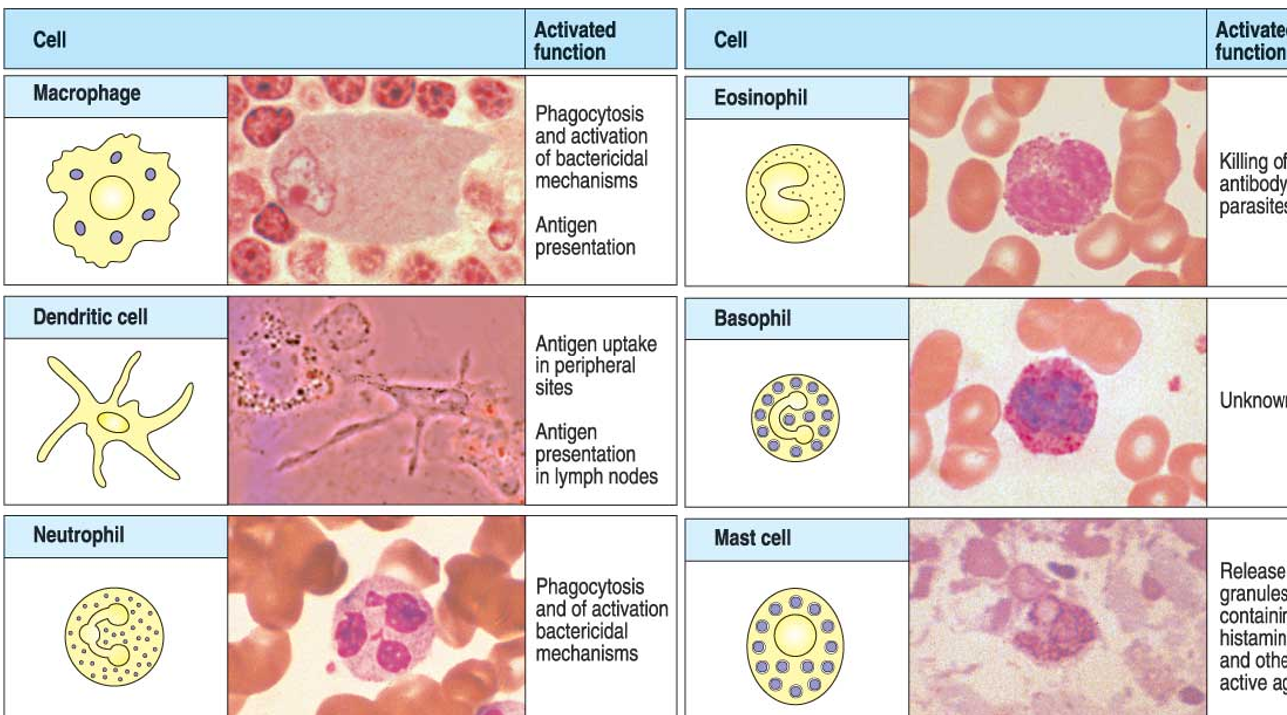
monocytes and macrophages
monocytes circulate in the blood
macrophages are formed by differentiation of monocytes nad are 5-10x bigger and found in tissues
macrophages ingest small pathogens and other material by phagocytosis
dendritic cells
present in most tissues and they have long cytoplasmic extensions (dendrites) to maximise antigen presentation to T cells and stimulation of adaptive immune response
granulocytes
neutrophils are phagocytic with lytic enzymes within granules including peroxidase and lysozyme: very effective in killing ingested bacteria
eosinophils are most important in defence against larger parasites
basophils are non-phagocytic and release active substances from their granules
NK cells
lymphoid lineage cells that are a part of the innate immune system
cytotoxic cells that kill virally infected/malignant cells: similar to T cells but without same activation requirements
cytotoxicity comes from pore forming molecules that are inserted into the target cell membrane and cytotoxic chemicals that enter target cell cytoplasm
PAMPs and PRRs
pathogen-associated molecule patterns (PAMPs) are molecular motifs commonly found in broad classes of pathogens and absent from humans
often glycoconjugates with the lipo-polysaccharides present in outer membrane of gram -ve bacteria being the prototypical example
pattern recognition receptors (PRRs) are present on innate immune cells (macrophages/dendritic cells)
recognise PAMPs and initiate a response
inflammatory response
recognition of pathogens by macrophages triggers signalling molecules (cytokines and chemokines) which together cause inflammation
blood vessels become more permeable and cause area to swell
leukocytes adhere to endothelial cells of blood vessels and pass between them to enter tissue
neutrophils are first to arrive, followed by monocytes, which differentiate into macrophages
inflammation is important in adaptive immunity as well: lymphocytes migrate into site of inflammation later
signalling and humoral response
cytokines are small protein signalling molecules of the immune system
e.g: interleukins, chemokines (induce chemotaxis) and interferons (released by virus-infected cells)
acute phase proteins: humoral factors that are upregulated or downregulated in response to inflammation
positive APPs: CRP and complement factors: both of which can function as opsonins, labelling microbes for phagocytosis
complement
part of innate immune system with links to adaptive immune system (ABs)
consists of 20+ different globular proteins found in blood plasma mostly produced by the liver
these proteins work in a cascade where one protein cleaves next to produce active fragments
3 pathways (classical, alternative, MBL) that converge with the cleavage of inactive C3 protein into active C3a and C3b fragments
result of cascade: to form membrane attack complexes which disrupt cell membranes of the pathogenic bacteria
classical pathway
ABs bound to antigens on bacterial cell surfaces bind the C1 complex
C1s cleaves C4, which binds bacterial surface, then cleaves C2
resulting split molecules form C4b2b enzyme complex (also called C3 convertase) which remains covalently bound to bacterial surface
C3 convertase cleaves C3 into C3a and C3b
C3b and its degradation products especially iC3b on pathogen surfaces, enhance phagocytosis
complex of C4b, C2a and C3b (termed C5 convertase) cleaves C5 into C5a and C5b
MAC assembles on target cells
alternative pathway
C3bBb can bind to another molecule of C3b and together they convert complement protein C5 into: C5a and C5b
C5b combines with complement proteins C6, C7 and C8 and forms a ‘stalk’ that anchors the protein complex into the bacterial cell wall
complement protein C9 completes the membrane attack complex (MAC), making a channel that opens a hole in the surface of the bacterium
MBL pathway
triggered by the soluble PRRs: Mannose Binding Lectin (MBL) and Ficolin: focuses on patterns not present on surface of human cells
MBL binds in the blood to a serine protease (MASP-1, MASP-2)
when MBL binds to terminal mannose sugars on microbial proteins, MASP functions like a convertase, cleaving C3 into C3b
C3b fragments then bind to bacterium and start the complement chain reaction