membrane bound organelles
1/18
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
19 Terms
animal cell
Cells are highly organised
Vast majority of proteins are produced on cytoplasmic ribosomes + encoded by nuclear genome
Others are produced in mitochondria
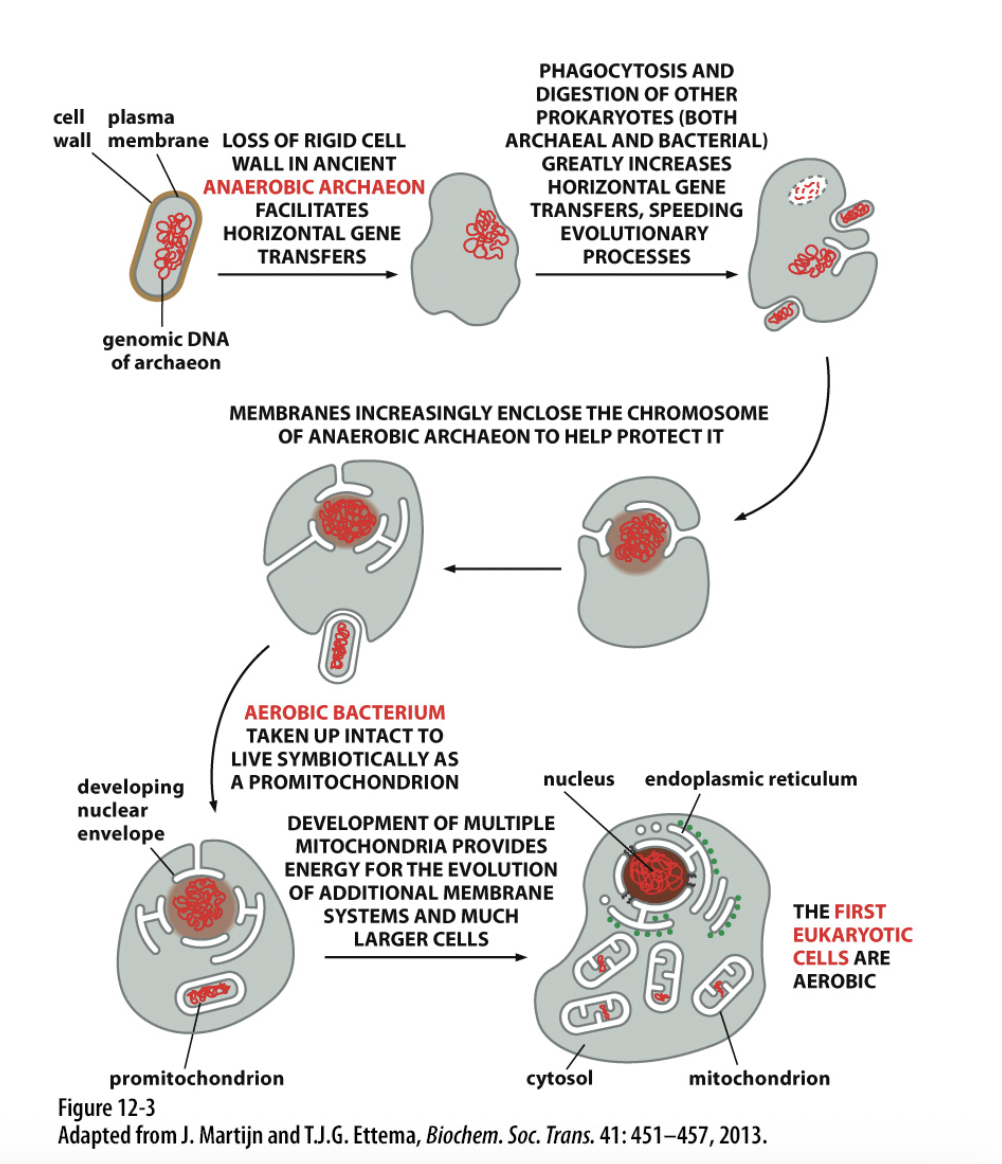
how do internal organelles evolve
Developed methods of invagination of the membrane
Thought to be some advantage of the way the membrane interacted with cell
Nuclear membrane formed first
Then endoplasmic reticulum
Are continuous with each other
Mitochondria and chloroplasts have a different origin
mitochondria, chloroplasts and peroxisomes
Mitochondria
Oxidative phosphorylation
Production of majority of cellular ATP
Chloroplasts
photosynthesis
Peroxisome
Fatty acid beta oxidation
Source of energy
endomembrane system
endoplasmic reticulum
rough
smooth
Golgi apparatus
vesicles
endoplasmic reticulum
Entry point of newly synthesised proteins
Translocon
Allows proteins to go into channel into endoplasmic reticulum
Doesn’t let ions in
Facilitates the assembly of integral proteins
If not they get into lumen of endoplasmic reticulum
Can stay or move onto other organelles
Integral proteins move laterally
Out of translocon into membrane
rough ER
Studded with ribosomes
Cytoplasmic ribosomes (on cytoplasmic side)
Dynamic
Interacts with system
smooth ER
No ribosomes
Involved in lipid synthesis
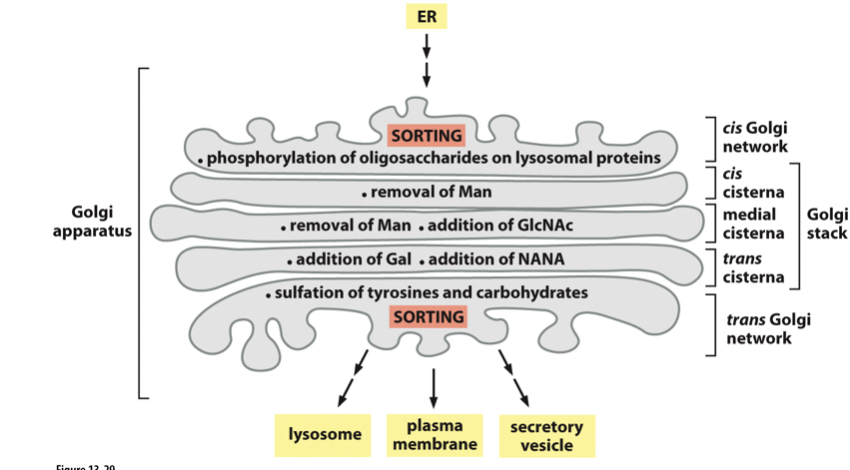
Golgi apparatus
Organised into Golgi sacs/cisternae
Cis face, trans face
New proteins travel from cis to trans
vesicles
Secretory
Release molecules to cell surface
exocytosis
Can be diverted
e.g. lysosomes
Form via budding
Fuse to release
why are there different compartments
Sequential processing / modification of secreted proteins
Important for these to happen in the correct order
Keep reactions separate
e.g. enzymes separate
processes that take place in the endoplasmic reticulum
N-glycosylation
Disulphide bond formation
folding and quality control
disulphide bond formation
Lumen = reducing site
So happens on lumen side
Proteins disulfide isomerase forms bond
Oxidation
Protein gets reduced
folding and quality control
Proteins not correctly folded do not leave ER
Role is not in ER, so will not start incorrectly functioning
Binds to glucosyl transferase (adds glucose)
Protein binds to calnexin
Keeps protein in endoplasmic reticulum
Sugar removed by glucosidase
If protein is still incorrectly folded then glucose added again
Once protein is correct, it will no longer be detected by glucosyl transferase and it can leave the ER lumen
Golgi apparatus
In stacks
Enzymes
Modify sugars on lipids and proteins
Cleave proteins (e.g proteases)
N and O linked glycosylation
N-linked
Adding sugar on asparagine
In ER
O-linked
Adding sugar on threonine/serine
ABO blood groups
Due to different sugar modifications
At antigen terminus
O= n/a
A = N-acetylgalactosamine
B=N-acetylglucosamine
N glycosylation
What it is:
Attachment of a sugar chain to the nitrogen (N) atom of the amide group of asparagine (Asn).
Where it occurs:
Begins in the rough endoplasmic reticulum (RER)
Further modified in the Golgi apparatus
Amino acid requirement:
Occurs at the consensus sequence:
Asn–X–Ser/Thr (X ≠ proline)
Process:
A pre-assembled oligosaccharide is transferred all at once to the protein.
The glycan is then trimmed and modified.
Functions:
Protein folding and stability
Cell–cell recognition
Protection from degradation
Examples:
Antibodies (IgG)
Many membrane and secreted proteinsO
O glycosylation
What it is:
Attachment of a sugar to the oxygen (O) atom of the hydroxyl group of serine or threonine.
Where it occurs:
Mainly in the Golgi apparatus
Amino acid requirement:
No strict consensus sequence
Process:
Sugars are added one at a time directly to the protein.
Functions:
Lubrication and protection (e.g., mucus)
Structural support
Cell signaling
Examples:
Mucins
Proteoglycans
lysosomes
Degradative organelles
First assembled in ER
Ph = 5 inside lumen (for enzymes)
Nucleases
Proteases
Glycosidases
Lipases
Phosphatases
Sulfatases
Phospholipases
Lumen reduced by a H+ pump
ATP -> ADP + pi
H+ pumped into lumen
protein sorting
A eukaryotic cell contains 10 billion (~1010) protein molecules
Cells are highly organised
Different organelles contain different sets of proteins.
If proteins were not sorted correctly chemical chaos would result.
Protein synthesis/degradation
Starts in the cytosol
4 outcomes
Stays put
Nucleus
ER trafficking
mitochondrion