M5 Van- RNA makes protein - translation
1/8
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
9 Terms
how is the amino acid code redundant?
multiple codons can code for one amino acid
in these cases, it is the third base in the codon that is unimportant and can be different
these are called ‘wobble bases’
normally the interchangeable bases will be the two purines or the two pyrimidines
in a few codons the third base can be anything
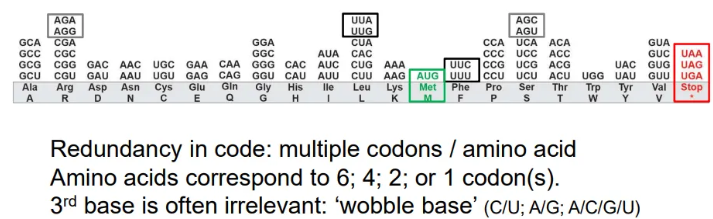
what are the start and stop codons in mRNA?
start
AUG
stop
UAA
UAG
UGA

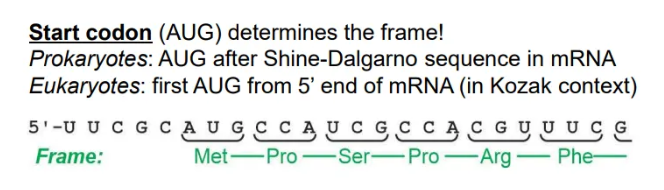
how is the correct open reading frame selected?
the first start codon (AUG) found will determine where translation starts, and hence the reading frame
since prokaryotic mRNA is polycistronic, this is the first AUG after the Shine-Dalgarno sequence
whereas in eukaryotes, this is just the first AUG from the 5’ end
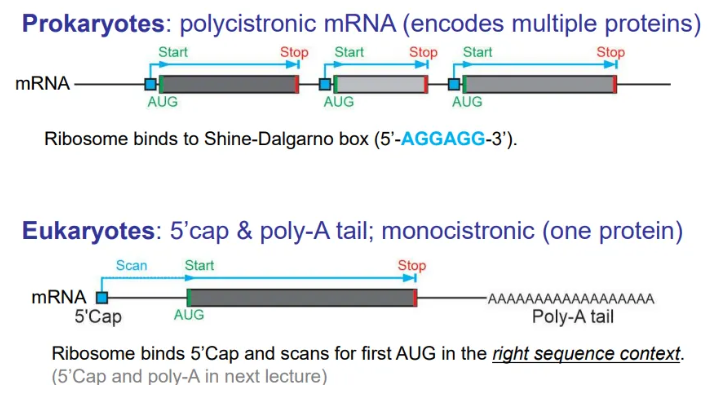
what are the general layouts of mRNA in prokaryotes and eukaryotes?
translation of prokaryotic mRNA begins before transcription is complete, because they don’t have membrane-bound nuclei, so the mRNA is polycistronic (encodes multiple proteins)
in eukaryotes the mRNA strands are cleaved and packaged to be sent to the ribosomes for translation, so the mRNA is in discrete chunks for each protein (monocistronic)
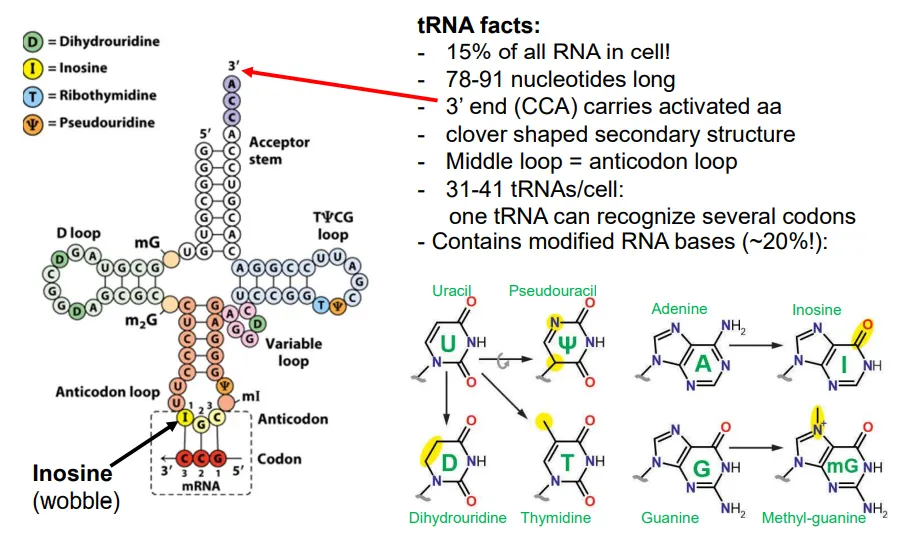
how do tRNA molecules deal with the redundancy of the genetic code?
there are only 31-41 distinct tRNAs per cell, compared to 60 codons, so many must have anticodons that can recognise the wobble bases
they do this by having their own modified RNA bases
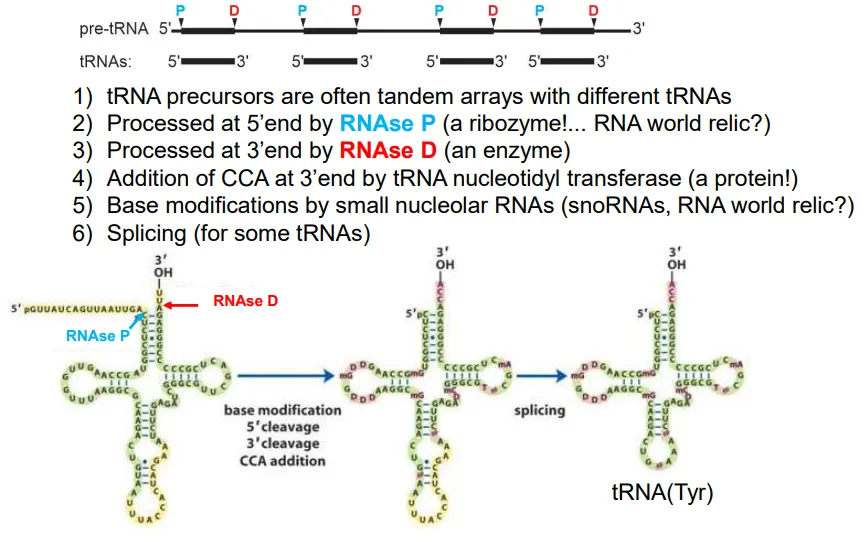
how are tRNA molecules produced?
the pre-tRNA molecules normally contain the sequences for multiple tRNAs (tandem arrays)
the sequence is cleaved at multiple points
at the 5’ end of each tRNA this is done by RNAse P, and at the 3’ end it is RNAse D
then the enzyme tRNA nucleotidyl transferase adds CCA to the 3’ end
base modifications occur in some tRNA molecules to deal with the wobble bases, and some get spliced
these molecules are then ‘charged’ by an aminoacyl-tRNA synthetase (aaRS) to add on the relevant amino acid to the new 3’ adenosine (at the 3’OH) using ATP
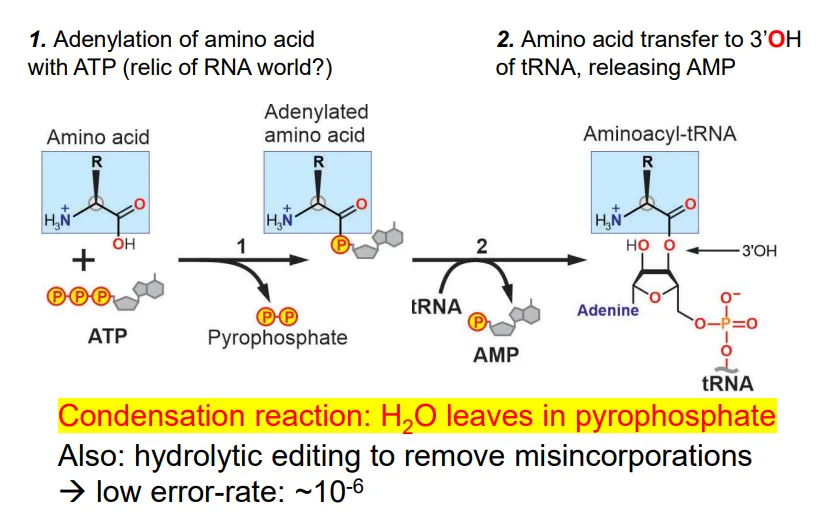
how are tRNA molecules charged?
the amino acid reacts with ATP to produce an adenylated amino acid (bound to AMP) and a pyrophosphate (contains water- this is a condensation reaction)
the AMP is replaced by a reaction with the 3’OH of the 3’ adenosine in the tRNA molecule
this reaction is catalysed by aminoacyl-tRNA synthetase enzymes (aaRS)- only 1 per amino acid)
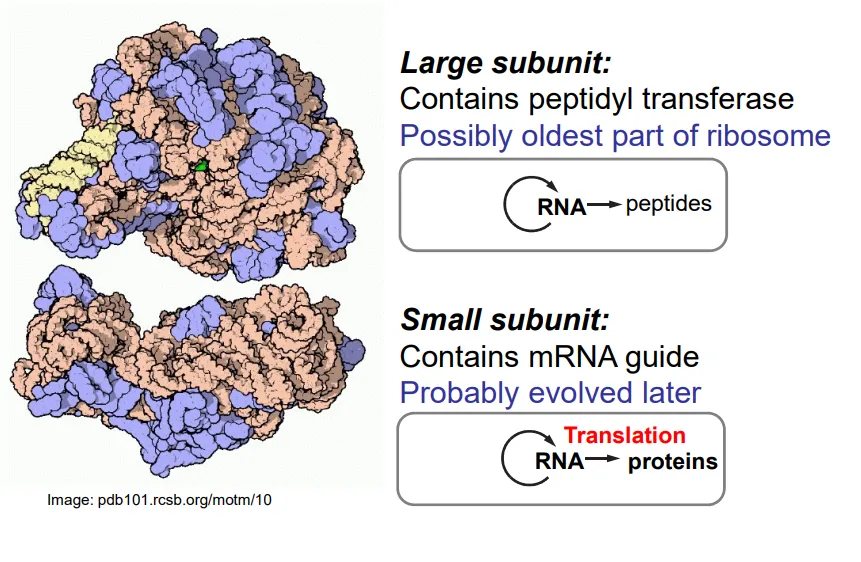
what is the structure of ribosomes?
ribosomes are mostly composed of RNA, with some peripheral auxiliary proteins
the large subunit contains peptidyl transferase (a ribozyme), which connects the amino acids by catalysing the formation of peptide bonds
the tRNA molecules bind to the large subunit
the small subunit binds to the mRNA so that codon-anticodon pairing can occur with the tRNA
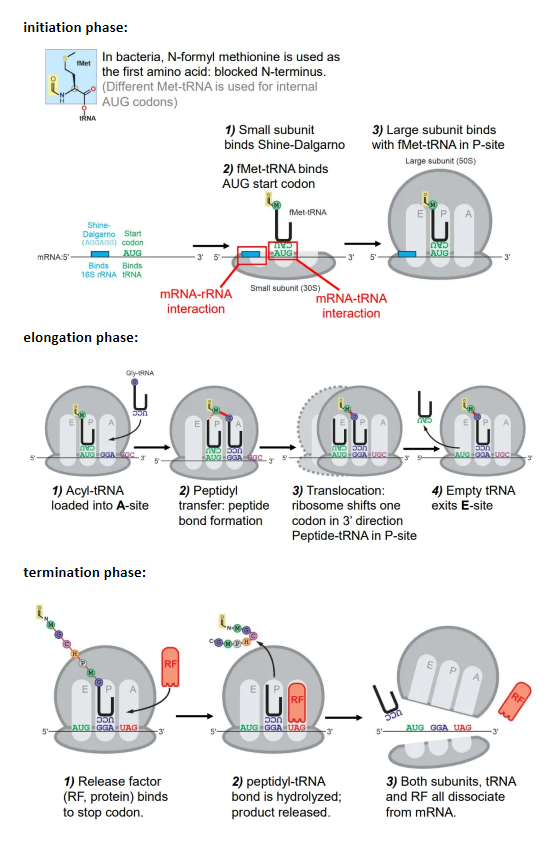
what happens in the initiation phase of translation in prokaryotes?
initiation:
the small subunit of the ribosome binds to the Shine-Dalgarno box
the Met-tRNA (methionine = start aa) binds to the first AUG codon after the Shine-Dalgarno box
the large subunit then binds to the Met-tRNA at the P site
elongation:
the charged tRNA molecule for the next codon is loaded into the A site, and a peptide bond forms between the two amino acids
the amino group of the incoming amino acid attacks the carbonyl carbon nucleophilically
this is catalysed by the peptidyl transferase ribozyme, which contains an adenine base that accepts and donates a proton to hydrolyse the peptide-tRNA bond
the ribosome shifts one codon, so the first tRNA is now in the E site, and exits
the second tRNA is now in the P site, so the process repeats, with new tRNA molecules being loaded into the A site and peptide bonds forming
termination:
a release factor (RF) protein binds to the stop codon at the A site
this causes the peptide-tRNA bond to be hydrolysed, so the peptide is released
both subunits, the tRNA and the RF protein dissociate from the mRNA