bio cycle 9 - phylogeny
1/36
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
37 Terms
what does a phylogeny (evolutionary tree) represent?
a hypothesis of evolutionary relationships showing branching patterns of descent from common ancestors, time flows from the root (most ancient) to the tips (most recent)
what is the most important feature when reading a phylogeny?
the sequence of branching events, not the left-right or top-bottom orientation
what is a node?
a branching point where a lineage splits; represents a speciation event and the MRCA of descendant taxa
what is the root of a phylogenetic tree?
the oldest common ancestor of all taxa in the tree; the point from which time flows toward the tips
what is a clade (monophyletic group)?
a group consisting of a common ancestor and all of its descendants; removable with a single “snip”
what is a non-monophyletic group?
any group that excludes some descents of its common ancestor or groups species that don’t share a recent common ancestor (paraphyletic)
what is a sister clade/sister taxa?
two lineages that share an immediate common ancestor; each other’s closest relatives
how can two different phylogenies represent the same information?
if the branching order is identical, rotating subtrees around nodes does not change evolutionary relationships
what is the taxonomic hierarchy (mnemonic: “Diligent Kindly Professors Cannot Often Fail Good Students”)?
domain → kingdom → phylum → class → order → family → genus → species
why do some traditional groups fail to reflect evolutionary relationships?
because many were defined by superficial similarity before evolution was understood; many (ex: fish, dicots, prokaryotes) are non-monophyletic
what is anagenesis?
evolutionary change within a lineage without branching
what is cladogenesis?
evolutionary branching that creates two or more descendant lineages
what is an ancestral character trait?
a trait evolved before the MRCA of the entire group
what is a derived character state?
a trait that evolved after the MRCA of the entire group; new in particular lineages
how do we determine ancestral vs derived states?
by comparing to an outgroup (a lineage that branched off before the ingroup
trait present in outgroup + all ingroup taxa?
ancestral
trait present in outgroup + some ingroup taxa?
ancestral
trait absent in outgroup but present in some ingroup taxa?
derived
trait present in outgroup but absent in all ingroup taxa?
equally likely ancestral or derived (because either the ingroup lost it or the outgroup independently gained it)
trait absent in outgroup but present in all ingroup taxa?
equally likely ancestral or derived (ingroup could have gained it or outgroup could have lost it)
what is a synapomorphy?
a shared, derived trait that evolved in the MRCA of only the taxa that have it, only type of characteristic useful for building phylogenies
what is a symplesiomorphy?
a shared, ancestral trait; present in the MRCA of the entire group, not useful for phylogenies because it does not indicate close relationships
what is an autapomorphy?
a unique, derived trait found in a single taxon; not useful for determining relatedness
why are synapomorphies preferred for inferring relationships?
because they indicate recent shared ancestry, taxa sharing more synapomorphies are more closely related
what is the principle of parsimony?
the simplest explanation requiring the fewest evolutionary changes is most likely correct
how is parsimony used in phylogenetics?
compare candidate trees; the one requiring the fewest gains/losses is the most parsimonious
ex: imagine four species, three of which can hover, and one that can’t
a parsimonious tree would suggest the hovering trait evolved once in the common ancestor of those three species, then was passed down, rather than evolving independently three separate times
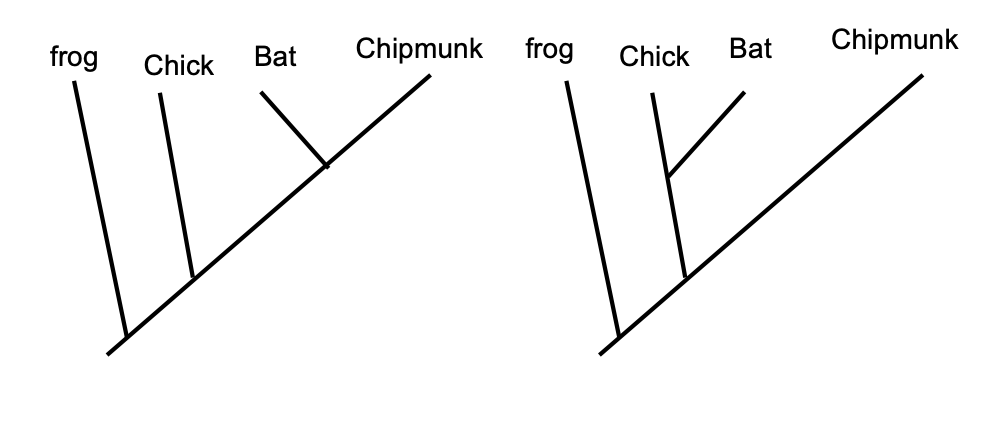
the chicken-bat-chipmunk example (outgroup = frog): which traits do each animal have?
outgroup: frog - no lactation, no wings, no fur, no beak
ingroup:
chicken - no lactation, wings, no fur, beak
bat - lactation, wings, fur, no beak
chipmunk - lactation, no wings, fur, no beak
are those traits ancestral or derived?
lactation - shared, derived
fur: shared and derived
beak - unique to chicken (autapomorphy/derived)
wings - shared, derived
no fur - shared, ancestral (outgroup and some of the ingroup lack fur)
which taxa share more synapomorphies?
bat and chipmunk share more derived traits (lactation + fur), so they are closer relatives than either is to the chicken
what is homology?
similarity due to shared ancestry; underlying structure is inherited from an ancestor even if function differs
what is convergent evolution?
independent evolution of similar traits in unrelated lineages due to similar ecological pressures; results in analogous structures (body parts in different species with similar functions but evolved independently from different ancestors and have different anatomy)
what is a homoplasy?
a misleading similarity or dissimilarity caused by convergence or reversal; does not reflect shared ancestry
why are homoplasies problematic in phylogenetics?
they mimic synapomorphies and can lead to incorrect trees
what are molecular clocks used for?
to estimate timing of divergence events by comparing approximate mutation rates
what is the comparative method?
using phylogenies to test hypotheses about trait evolution while accounting for shared ancestry
what is the cladistic method of classification?
only monophyletic groups (clades) receive names; classification must reflect evolutionary history
steps for building a phylogenetic tree (parsimony approach)
choose an outgroup
compare traits across taxa
identify ancestral vs derived traits
group taxa by shared derived traits (synapomorphies)
select the tree with the fewest evolutionary changes