Lecture 18: Transmembrane proteins, Golgi, COP
1/16
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
17 Terms
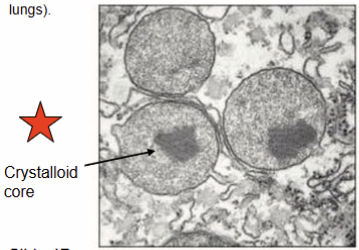
Peroxisome
type of organelle called a microbody
found in almost all eukaryotic cells
crystalloid core: contains enzymes needed for the reaction
involved in some enzymatic reactions
e.g. catabolism of very long fatty acid chains
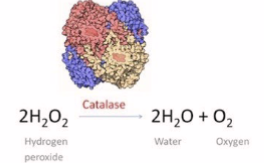
reduction of reactive oxygen species (photo)
biosynthesis plasmalogens
phospholipids for the function of brians and lungs
Zellweger syndrome
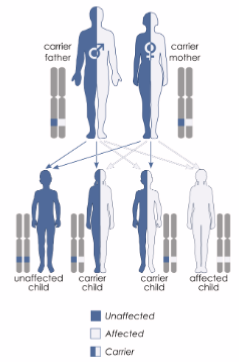
inherited in an autosomal recessive
severe brain development defects
Hypomyelination
Apnea (stop breathing)
Abnormal renal function
Patient usually does not survive beyond one year
Cystic fibrosis
caused by mutation in gene, cystic fibrosis transmembrane conductance regulator (CFTR)
the most common mutation, ΔF508, is a deletion of three nucleotides, resulting in a loss of the amino acid phenylalanine (F) at the 508th position on the protein. This mutation accounts for two-thirds (66–70%) of CF cases worldwide

Cotranslational protein import
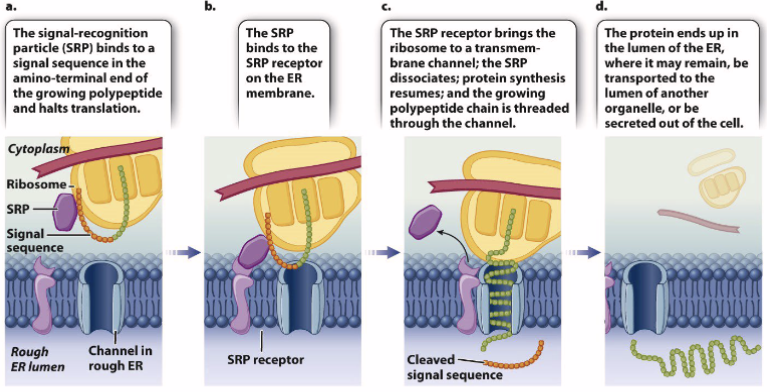
How transmembrane proteins are integrated into a membrane
type 1 single-pass transmembrane proteins
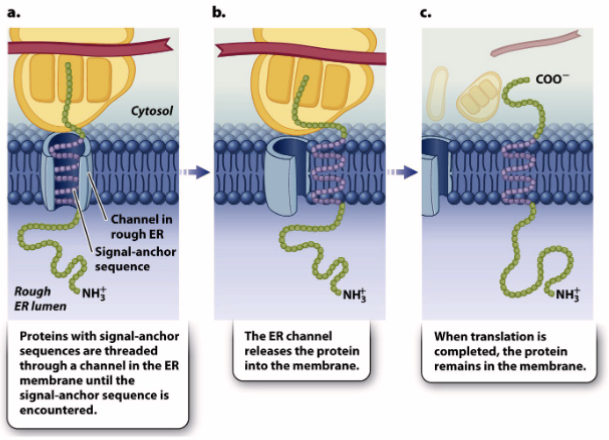
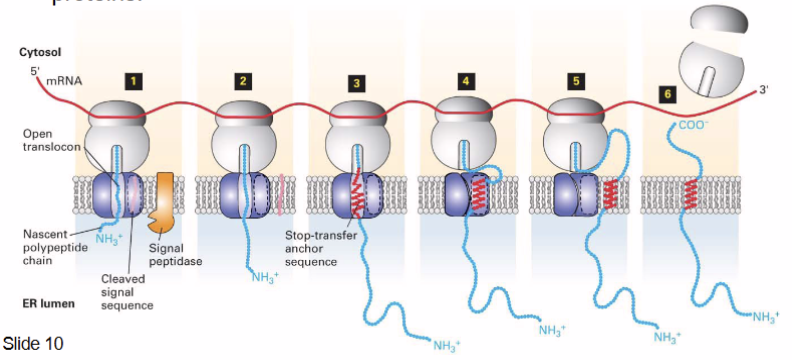
synthesis of integral membrane proteins
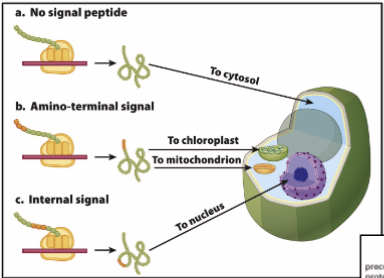
How proteins are targeted to mitochondria and chloroplast
N-terminal sequences direct proteins to their respective organelles, which are eventually directed to the right compartment/membrane
mitochondria: OMM, IMM, intramembrane space, matric
chloroplast: OCM, ICM, thylakoid membrane/lumen, stroma
endocytic pathways for protein sorting googoo gaagaa
after a protein is fully synthesized, folded, and targeted to the ER lumen, it either:
stays in the ER lumen
transported from ER → Golgi complex → secretory pathway
fully processed proteins are exported to TGN
transfer of ER vesicle → Golgi is achieved by coat proteins (helps form vesicle, helps select material vesicle carries)
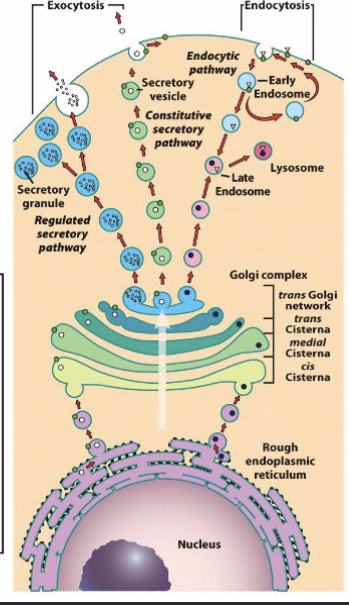
ER to Golgi complex
Golgi apparatus receives proteins and lipids from the ER and to other organelles, plasma membrane, or the cell exterior; proximal → distal; Cis-Golgi Network (CGN) → medial Golgi → Trans-Golgi Network (TGN)
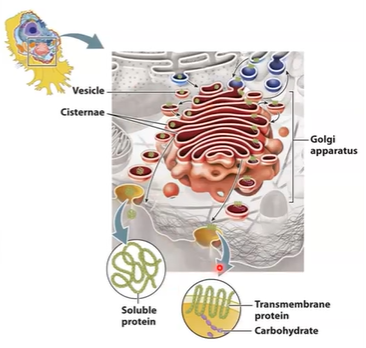
Structure of Golgi complex
smooth, flattened disk-like cisternae
~8 cisternae/stack - several 1000s
cisternae are biochemically unique
curved like shallow bowl
shows polarity
Membrane supported by protein skeleton (actin, spectrin)
Scaffold linked to motor proteins that direct movement of vesicles into and out of the Golgi.
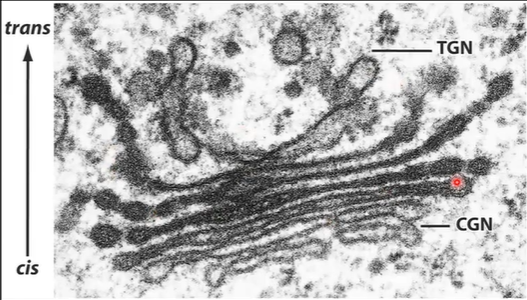
Difference between Golgi complex parts
CGN acts as a sorting station (i.e., sorts whether proteins should
continue on to the next Golgi station or be shipped back to the ER)TGN sorts protein into different types of vesicles—vesicles go to plasma membrane or other intracellular destinations (e.g. lysosomes)
biochemical diversity of Golgi complex
Proteins are modified step-wise as they traverse the Golgi.
Different cisternae of the Golgi contain different enzymes that modify proteins.
The differential staining of the Golgi cisternae reflects their biochemical difference
processing plant of the cell
involved in synthesis of polysaccharides and specific modification of protein sna dlipids (glycosylation and proteolytic modification)
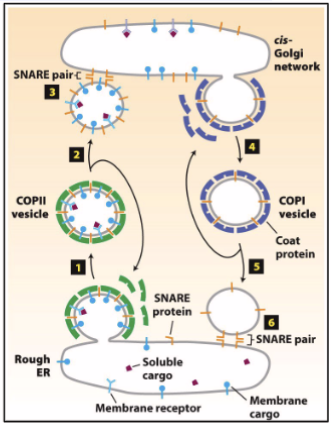
Coatomer; COPI and COPII
Coat protein complex (COP) COPI and COPII are protein complexes that assemble on the cytosolic surface of donor compartment membranes at sites where budding takes place; electron micrographs reveal COPI and COPII on vesicles
COPI in retrograde direction (opposite directions)
COPII in anterograde direction
Key features of lysosomes
Digestive organelles.
Size: 25 nm to 1 μm.
Internal pH of 4.6 (proton pump or H + -ATPase). Contains hydrolytic enzymes: acid hydrolases.
Lysosomal membrane is composed of glycosylated proteins that act as a protective lining next to acidic lumen
Function of lysosomes
Autophagy: normal assembly of unnecessary/dysfunctional cellular components
Degradation of internalized material
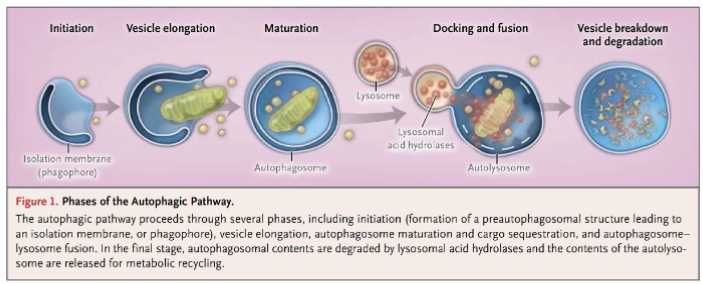
Autophagy
Autophagosome formation → lysosome recruitment → autolysosome → digestion and release by exocytosis
decomposition of intracellular components via lysosomes
plays an important role in maintaining/regulating homeostasis by degrading components and providing degraded products
Isolation membrane derived from ER engulf target organelles to form an autophagosome (also known as autophagic vesicle)
Lysosome fuses with ER-derived autophagic vesicle to form an autolysosome.
Content of autolysosome is enzymatically digested and released (exocytosis)
Degradation
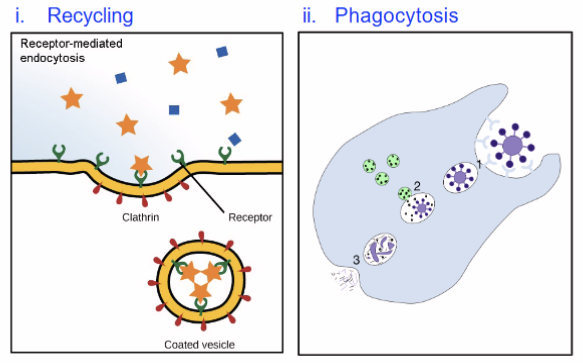
Recycling of plasma membrane components like receptors and extracellular material
Destroy pathogens like bacteria and viruses—only in phagocytic cells