L9-15: Macroevolution
1/109
Earn XP
Description and Tags
L9 + 10 Speciation,L11 Phylogenetics L12 Molecular Phylogenetics L13 Molecular Evolution L14 Gene Family Evolution L15 Genome Evolution
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
110 Terms
What is the essentialist concept of species?
a set of pre-defined or created qualities
views species as immutable (new species cannot arise).
This became untenable with Darwin and the theory of evolution
What is the morphological species concept?
A similarity-based concept that defines species based on APPEARANCE
subjective
useless for polymorphic species
misleading for species that look the same but not (cryptic species)
Define the Biological Species Concept (BSC)
Species are groups of actually/potentially interbreeding natural populations
also REPRODUCTIVELY ISOLATED from other species groups (Reproductive Isolation) [RI]
(Ernst Mayr, 1942)
What is Pre-zygotic Isolation?
RI that gametes do not come together
4 isolation mechanisms:
geographic (physical iso.)
ecological (physical iso.)
behavioral (NO physical iso.) e.g. mating activity
temporal (NO physical iso.)
What is Post-zygotic Isolation?
RI where gametes meet, but no (or sterile) hybrids are produced
Hybrid zygote fails to develop.
Hybrids are unfit or sterile (e.g., Horse x Donkey = Mule)
What are Ring Species?
A series of populations that are hybridizing sub-species connecting two ends.
At the "ends": populations exhibit
complete Reproductive Isolation (RI)
do not breed (e.g., Ensatina salamanders or Phylloscopus warblers)
What is the Phylogenetic Species Concept (PSC)?
A historical-based concept
defines a species as "an irreducible (basal) cluster of organisms that is diagnostically distinct from other such clusters
→ within which there is a parental pattern of ancestry and descent.
Drawback: deciding where to "draw the line" on the phylogenetic tree
What is Allopatric Speciation?
(Isolated) Speciation where populations are ISOLATED by a physical barrier.
Initial step: Barrier formation prevents gene flow.
Process: Populations diverge due to genetic drift and local selection.
Result: If the barrier is removed, Reproductive Isolation (RI) has evolved.
What is Peripatric Speciation?
(Isolated) Speciation involving an ISOLATED NICHE.
Initial step: A subset of individuals enters a new, nearby niche.
Process: Populations diverge via genetic drift (especially "Founder effect") and inbreeding in the new habitat.
Example: Origin of Polar Bears from Irish Brown Bears.
What is Parapatric Speciation?
(Near) Speciation involving a NEARBY NICHE.
Initial step: A new niche is entered adjacent to the original range.
Process: Typically occurs where the environment abruptly changes. Hybrids fall into an "adaptive valley" (unfit), leading to selection against hybridization.
Example: Anthoxanthum odoratum grass on contaminated vs. uncontaminated soil.
What is Sympatric Speciation?
(Together) Speciation that is NOT ISOLATED (occurs within the population).
Initial step: Genetic polymorphism arises.
Process: Disruptive selection leads to the formation of local 'races' or preferences (e.g., insects preferring different host plants). Known as "Speciation with gene flow."
Example: Rhagoletis pomonella (apple maggot) diverging into apple- and hawthorn-feeding races.
What is the Bateson-Dobzhansky-Muller (BDM) Model?
A model explaining how genetic incompatibility evolves.
Two populations diverge in isolation (e.g., AABB becomes AAbb and aaBB).
Allele 'a' and allele 'b' have never been in the same individual.
If they hybridize, the interaction between 'a' and 'b' (AaBb) may be incompatible/unfit.
What is Reinforcement?
The process where natural selection strengthens reproductive isolation.
If hybrids are less fit (partly adapted),
selection favors non-hybrid matings (e.g., divergent flowering times or life cycles) to prevent the production of unfit offspring.
What is Polyploid Speciation?
A form of hybrid speciation involving an increase in chromosome number (e.g., 2x + 2x = 4x).
Occurs when unreduced gametes (diploid) fuse.
Common in plants (15-30% of speciation events).
What is Homoploid Hybrid Speciation?
Hybrid speciation without an increase in chromosome number. The diploid hybrid gains reproductive isolation, most likely due to ecology (novel ecological tolerances).
Example: Hybrid sunflower Helianthus paradoxus (transgressive segregation)
What is Systematics?
the methods and practices used to describe, name and classify biological diversity
What is Classification?
the result of systematic investigations
What is Taxon (pl.taxa)?
a unit in systematics, a group at any level in the systematic hierarchy (often used in place of ‘species’)
What are Characters?
the groups of attributes (qualitative or quantitative) used to define a species (e.g. morphology or DNA sequences)
The Linnaeean Hierachy
Hierarchical structure (species within genera, within families, within orders etc.)
What are Morphological systematics
Using morphological, anatomical, behavioural data to infer phylogenetic history
Principle of parsimony is most often used
What is Parismony?
PHYLOGENETIC RELATIONSHIP
simplest tree possible
(if looks the same → sort into a group)
What is Occam’s (Ockham’s) razor?
among competing hypotheses
the hypothesis with the fewest assumptions should be selected (MOST PARSIMONIOUS)
most parsimonious representation of data = FEWEST CHANGES EACH BRANCH
What are Phylogenetic trees?
A graph with internal and external nodes connected by internal and external branches
Branch length DON’T represent distance (simple trees)
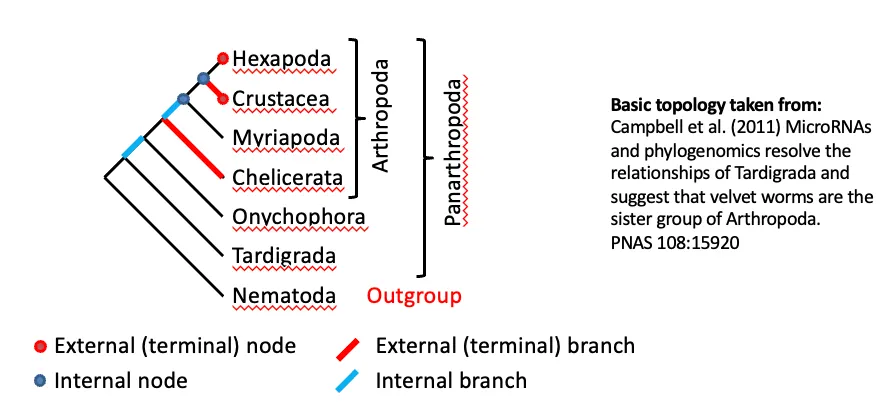
Bifurcating trees v.s. Multifurcating trees
Bi: 2 branches from it and FULLY RESOLVED
Multi: have POLYTOMY (don’t know relationships of 2 groups)
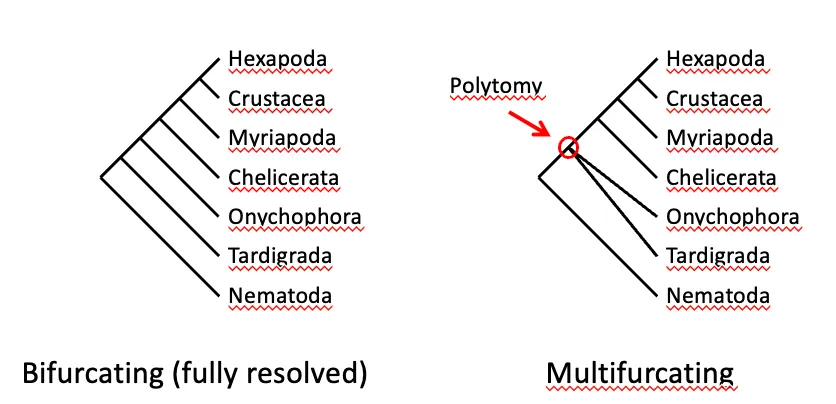
What is a outgroup?
defined by our own (not from the phylogenetic tree)
at BOTTOM LINE of tree
e.g. round tree and branches tree example
What is Strict Parsimony?
used when there >1 parsimonious trees
BUT lose of a lot of info if lots of trees
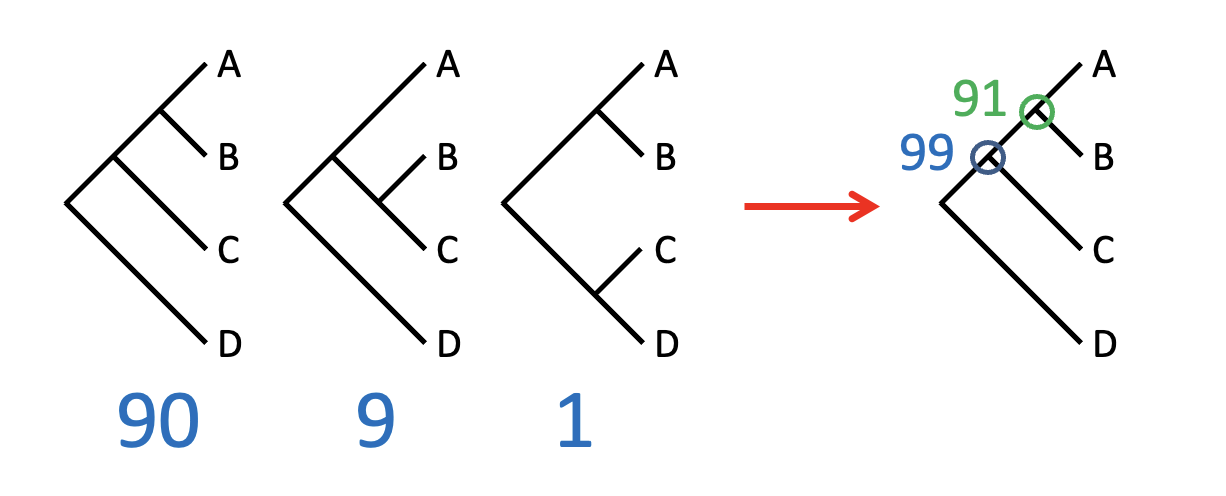
What is and how to use boot-strapping: Improving strict parsimony
What? : assessing support for phylogenetic relationships
How: Resampling the data
remove char one at a time
recalculate tree
see how many times different topologies are resolved
0 – most likely
91 % of time a and b are more mostly related
99% - of time A+B and C are more related
70% fine – 90% good and so one
→Tell how robust it is
What is Molecular systematics?
The use of DNA or protein sequences to infer phylogenetic history
Several methods of analysis can be applied
What are morphological characters used for? and downsides
can be useful in determining morphological relationships
might not always be representative of the evolutionary history of the species
What is Homoplasy?
Traits evolve more than once
not by DNA and protein sequences
e.g. wings in birds, bats, and flower colour
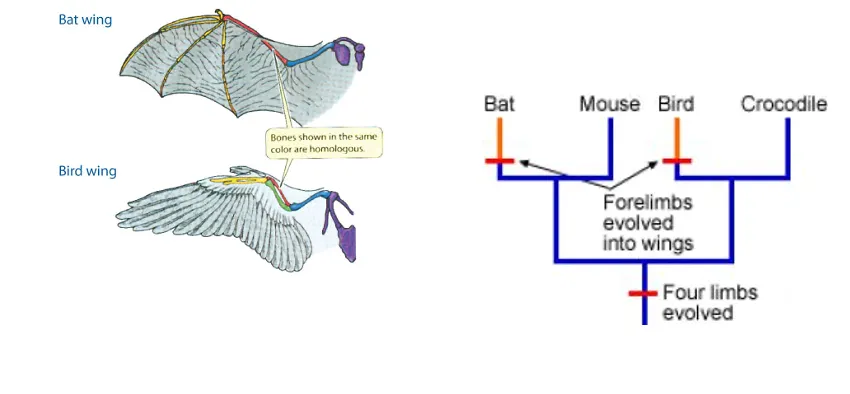
Why are DNA sequences more reliable than morphology of phylogenetic reconstruction?
less likely to show homoplasy
objective
Slow-evolving sequences v.s. Fast-evolving sequences
Slow:
too similar to be informative when looking at closely-related species
for big groups e.g. family of plants
Nuclear gene picked
Fast:
too dissimilar to be informative when looking at distantly related species
Mitochondria gene picked
Choose slow/fast evolving sequence
depending what taxa interested
Where are phylogenetics used in biological disciplines?
Forensics and fossilised DNA
Origin of species, origin of a characteristic
Biogeography and the history of the earth
Conservation and extinctions
Crop science and feeding the world
Making phylogenetic tree
using characterized DNA sequence (ACTG)
based on similarity of DNA sequence (min. no. of changes along tree
not simple as all DNA substitutions are NOT EQUAL
DNA - main takeaways
ALL DNA nucleotides CAN MUTATE (lost elimiated by natural selection)
TRANSITION MORE COMMON than transversions
(A,G = purines, C,T = pyrimidines)
some Mutations change a.a sequence - minor or major consequence
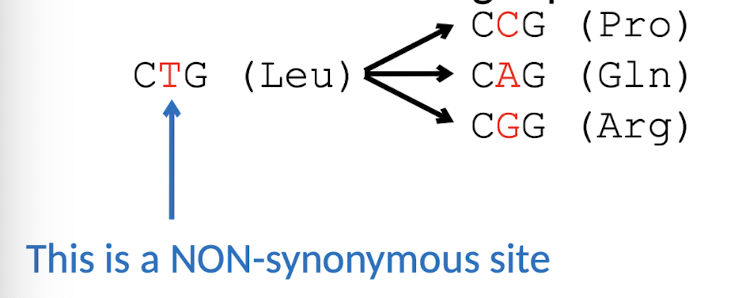
Synonymous v.s. Non-synonymous subsitutions
No change in AAs (LIKELY sub) → Change in AAs (UNLIKELY sub)
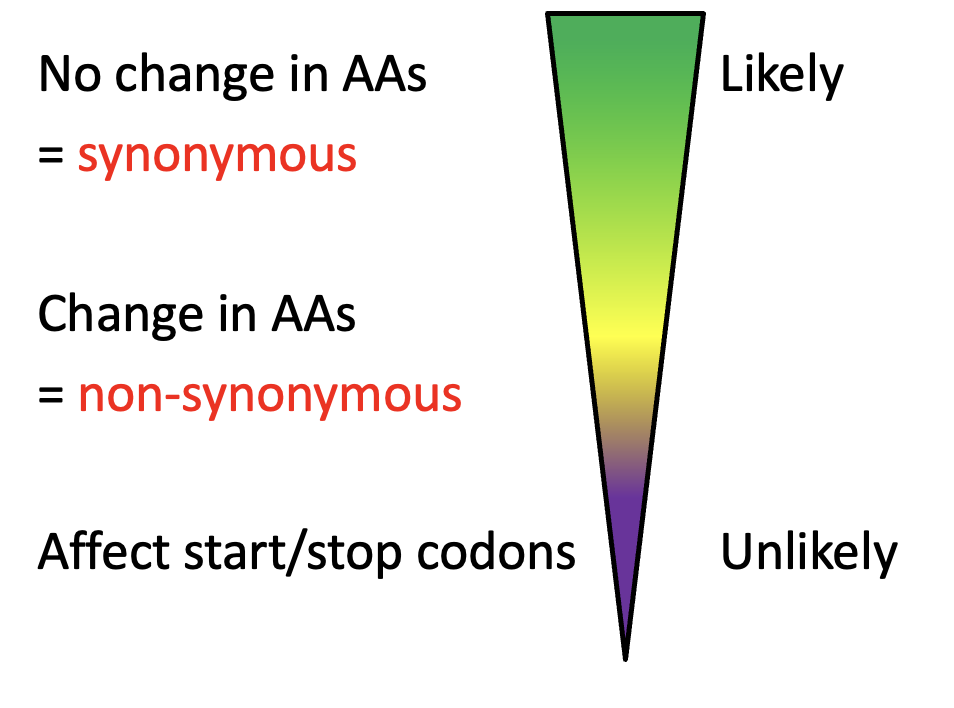
→ affect start/stop codons
What does gene trees and species tree show? and what is proven by?
Allele at all genes in an organisms DON’T have the same history as the organism
Proved by:
Sharing of ancestral variation - e.g. gorilla and orang-utan
cp/mtDNA introgression - hybridisation of species
Avoid errors in phylogenetic analysis (x rely on one gene) by:
Carry out different types of phylogenetic analysis
Analyse more than one gene (individually or in combination)
Sequence more than one individual per species
What cause inaccurate phylogenies?
Hybridization
Sharing of alleles
2 types of tree-drawing methods
Distance:
Using aligned DNA or amino acid sequences calculate the number of differences between all pairs of sequences
Computational
Max. parsimony:
use UPGMA
check ppt notes
draw in scale

What can phylogenetics tell us with ancient DNA?
relationships between modern humans and Neanderthals
relationships between mammoths, dodos etc. and modern-day species
the diet of early humans: e.g.
DNA taken from the stomach of a 5,200-year-old frozen human (Ötzi, the Tyrolean Iceman)
His genome tells us he was from Sardinialy humans
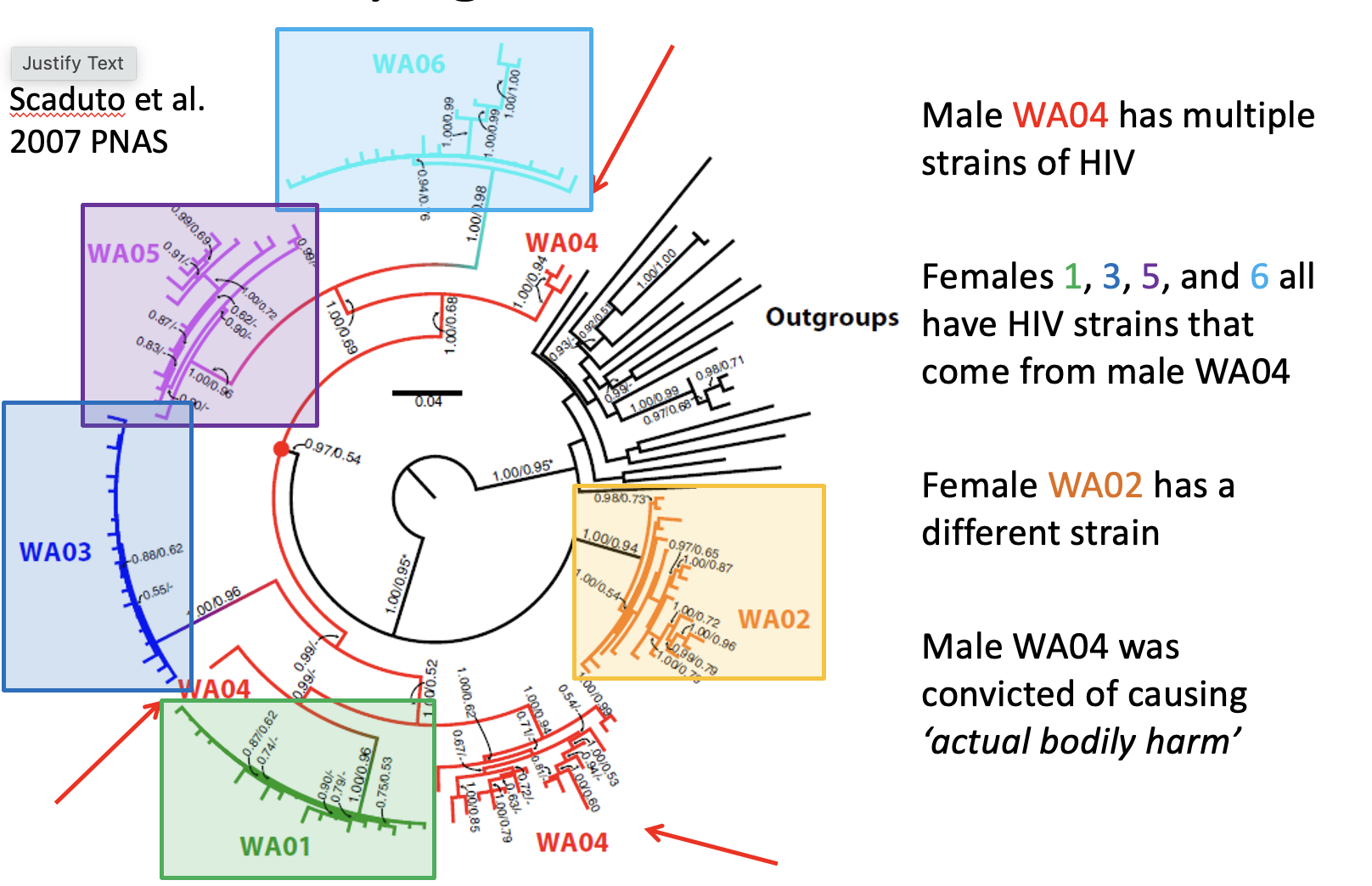
What can phylogenetics tell us in forensics?
track transmission of HIV
2 cases in the USA
In each case, based on phylogenetics of HIV samples → the source of HIV in multiple infected females was tracked back to one male (Scaduto et al. 2007)
What can phylogenetics tell us about diseases?
Track diseases → see mutation required
e.g.
H7N9 (‘bird flu’)
jump from infecting domestic chickens and ducks to infecting people at the end of March 2013
Phylogenetics reveals the closest related viruses were from ducks (Liu et al. 2013)
COVID
shows multiple sources
How can phylogenetics help with conservation?
Clouded leopard, SE Asia: Identifying endangered species for conservation
-similar genetic distance between major groups and between lions and tigers → consider the clouded leopard to be two species
implicates conservation of endangered species
Ivory trade: track source of poaching
Sequenced 7 genes from 574 elephants
Sequenced genes from tusks being illegally traded
Poaching is clearly concentrated in/near Zambia
→ combat poaching
Parrot trading (yellow-faced parrot): stop illegal trading of endangered species
DNA from illegally traded eggs, skin, feathers etc. can be used to identify the species and whether it is on the CITES list
DNA testing of 50 birds eggs strapped to a passenger at an airport in Brazil
What three population-level processes primarily determine the rate of molecular evolution?
Mutation rate (μ): The frequency at which new mutations occur.
Population size: Impacts genetic drift and the fixation rate of alleles.
Selection: The process favoring advantageous traits or removing deleterious ones
What are Mutations?
Stochastic permanent physical changes of DNA
by errors in DNA replication
often are changes in a single base (could be large scale)
How does population size influence the fixation rate of alleles?
Small populations: Alleles fix faster due to the sampling effect (genetic drift)
Large populations: There are more mutations overall,
→ but the fixation rate of a specific neutral allele is proportional to its allele frequency
When are there MORE MUTATIONS?
LARGER POPULATIONS
SHORTER GENERATION TIME
What are the specifics of the human mutation rate?
Human mutation rate (μ) :
approx. 2.5 × 10⁻⁸ substitutions per site per generation
3.2 GB genome size
→ results in roughly 13 DNA changes per individual per generation
Relationship between fixation rate (neutral allele) and allele frequency
they are PROPORTIONAL
Difference in allele outcomes between
100 vs.10 population after 50 generations?
100 Individuals: Some alleles are lost, but most are still segregating (more mutations)
10 Individuals: Many alleles are lost completely, and very few are still segregating (inbreeding depression)
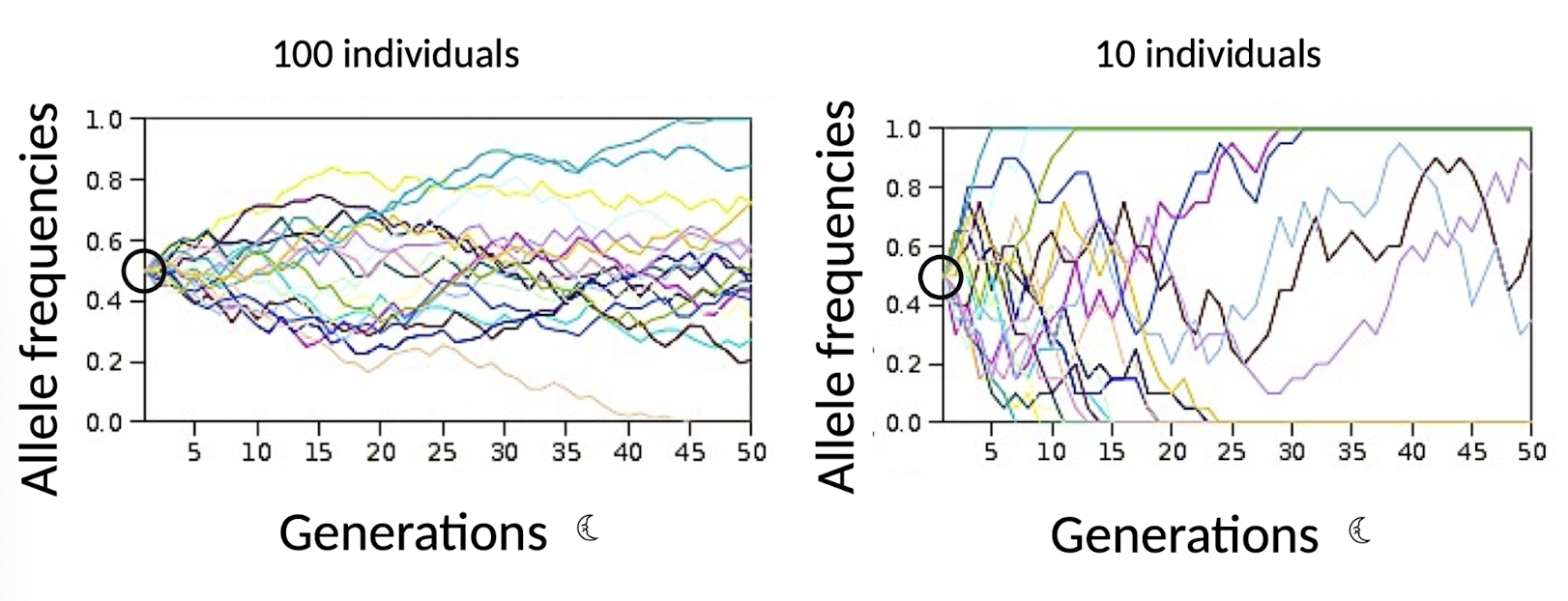
What are small populations (10 indivduals) prone to?
INBREEDING depression
3 types of mutations
Mutations are
Detrimental (negative)
Neither detrimental nor beneficial (neutral)
Beneficial (positive)
e.g. in a novel environment the mutation causes the encoded protein to function better in a new environment
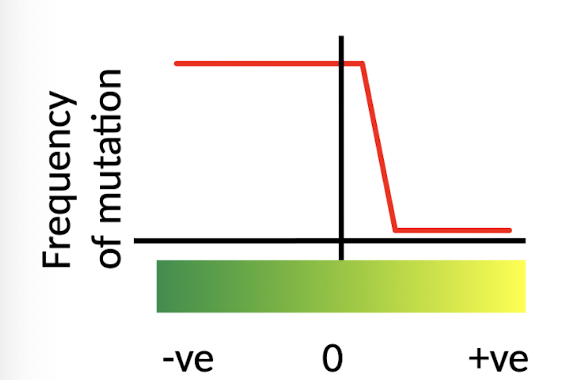
What does Kimura’s (1968) Netural Theory of Evolution propose?
POSITIVE selection -RARE
NEGATIVE mutations - removed by natural selection
Most mutations are neutral → molecular evolution governed by GENETIC DRIFT
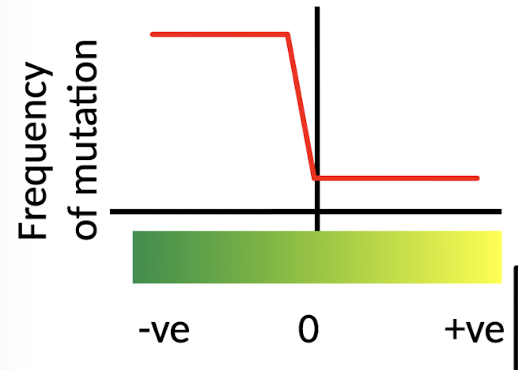
What does ‘Selectionist’ Theory of Evolution propose?
True neutrality - RARE
Negative mutations - removed by natural selection
Most mutations are NOT neutral → molecular evolution governed by SELECTION
Detecting POSITIVE selection: What does dN (or Ka) and dS (or Ks) represent?
dN (or Ka) represent:
number of non-synonymous changes (Dn) per non-synonymous site
Are mutations that result in a different amino acid
few ADAPTIVE mutations
dS (or Ks) represent:
number of synonymous changes (Ds) per synonymous site
are mutations that result in the same amino acid
most NEUTRAL mutatons
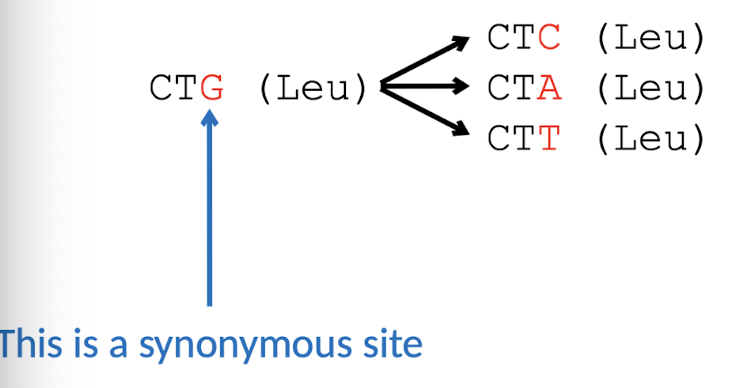
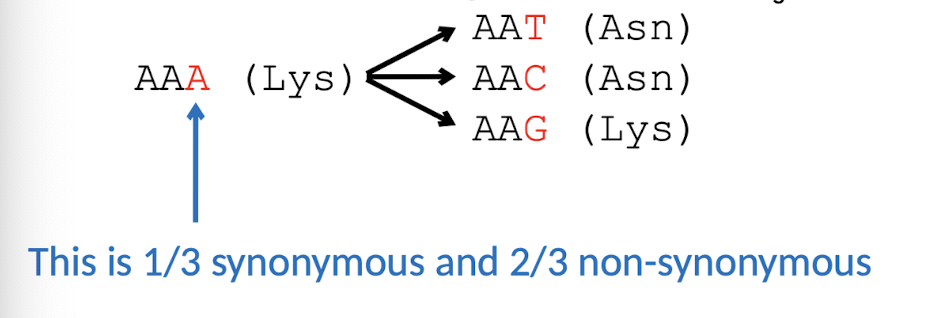
Average gene ratio of dN/dS
dN/dS < 1
3 different ratio of dN/dS indicates (selection-wise):
dN /dS < 1 → selection AGAINST change (few NS changes)
dN /dS = 1 → neutrality (equal likelihood of NS and S changes)
dN /dS > 1 → selection FOR change (many NS changes)
What is the "Red Queen Hypothesis"?
a species must "run faster and faster in order to keep still where she is."
explain co-evolutionary arms race (between host & parasites)
How does the Red Queen Hypothesis apply to host-parasite interactions?
Parasite numbers increase as hosts increase
→ negative effect on the host, numbers decrease
Hosts evolve immunity, numbers increase
→ parasites then evolve to overcome that immunity
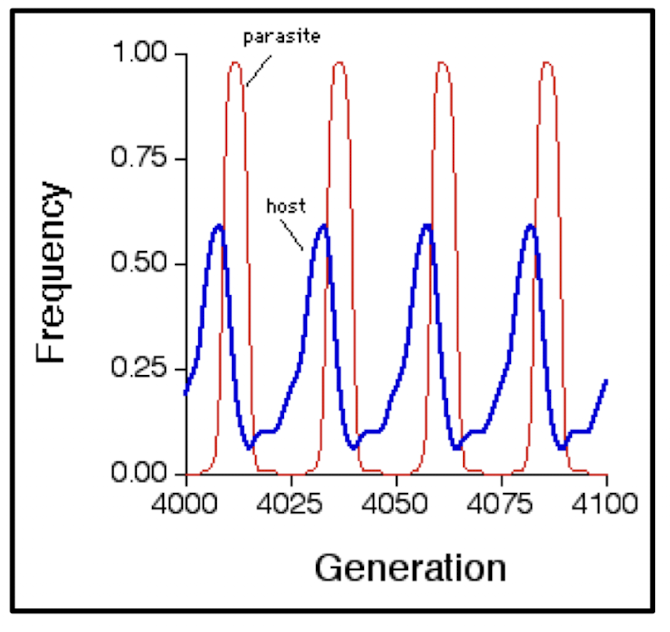
How does Red Queen Hypothesis occur in immune-related (virulence-related) genes ? and how is ideal to detect positive selection?
very strong selection between host and parasite
No immunity = DEATH
have mutation = DON’T DIE (greater fitness advantage)
What drives the positive selection (dN/dS > 1) of
Lysin (sperm-egg recognition) genes in Abalone?
Speciation and reproductive isolation
Abalone are broadcast spawners
1:1 recognition between sperm lysin and the egg's vitelline envelope for coexistence of species
As new species evolve, lysin must evolve rapidly to prevent cross-species fertilization
What is a Selective sweep?
when VERY STRONG SELECTION will remove ALL alleles but keep FITTEST
gene swept of diversity
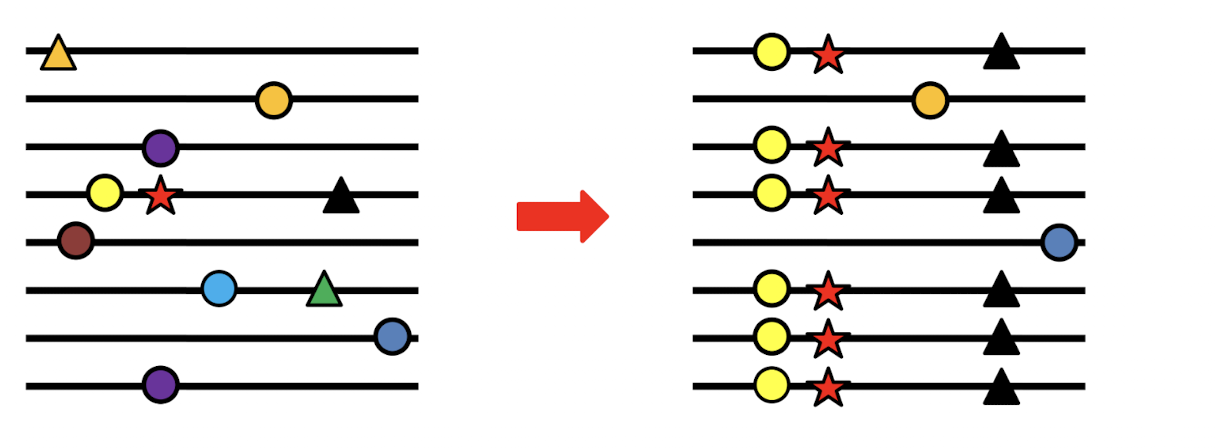
How do we detect a selective sweep in genomic data?
look across individuals for a "signature of selection"
specifically regions where a gene has very low diversity compared to the rest of the genome.
Testing selection with dn/ds or selective sweeps:
What genomic evidence of speciation was found in Anthoxanthum odoratum plants molecularly adapted to contaminated soil?
Parapatric speciaton:
Specific alleles for poor soil were found at
very high frequencies (e.g., 0.98 or 1.00) in the serpentine (contaminated) population
rare or absent (0.00) in the granitic population
Testing selection with dn/ds or selective sweeps:
What gene is responsible for molecular adaptation to hypoxia (low oxygen) in Tibetan humans, dogs, and yaks?
EPAS1 (Wang et al. 2014 GBE)
involved in the response to hypoxia, including angiogenesis and energy metabolism
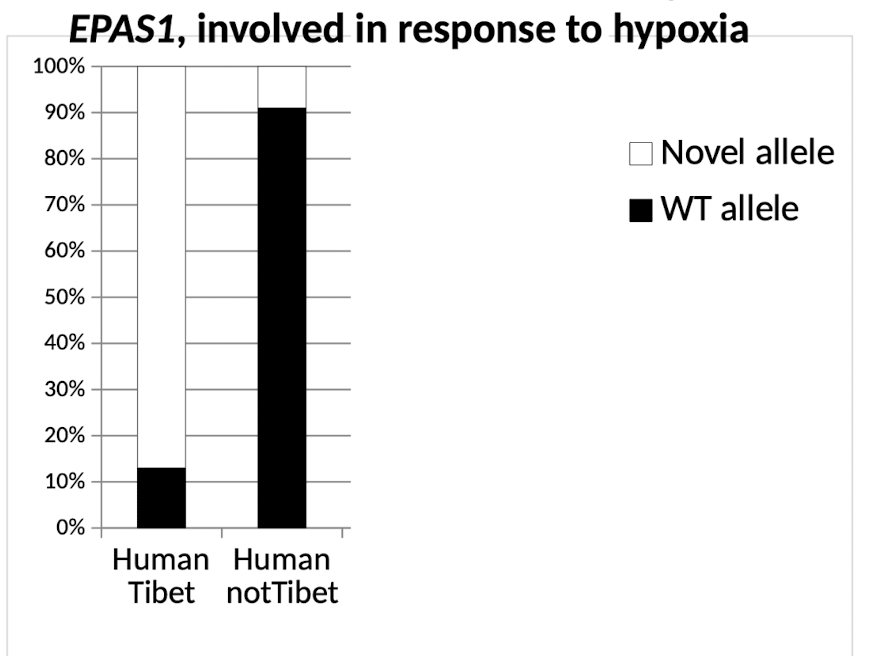
Testing selection with dn/ds or selective sweeps:
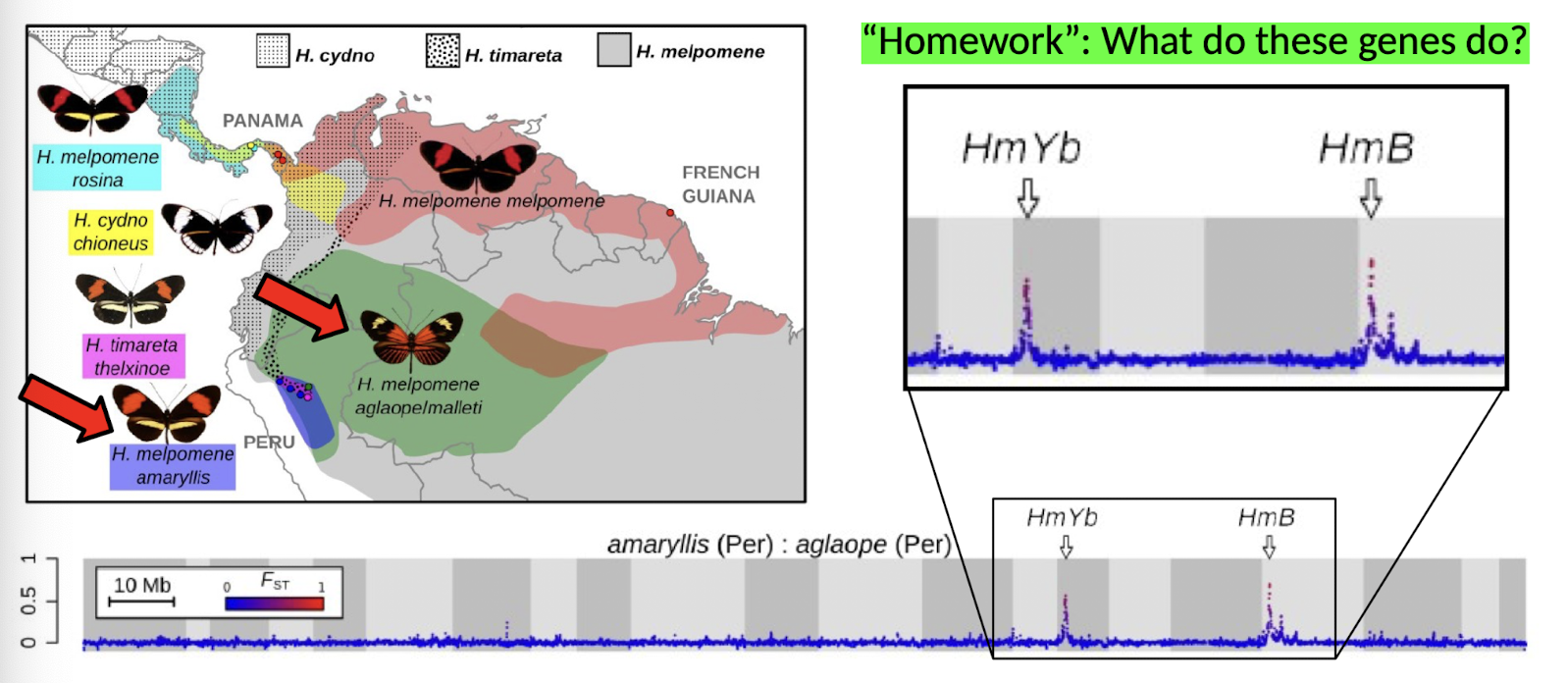
What is the genetic basis for wing pattern mimicry in Heliconius butterflies?
The genes HmYb and HmB (Martin et al. 2012)
Genomic scans show high differentiation (peaks in FST) at these loci
→ control wing patterns shared between species like H. melpomene and H. cydno
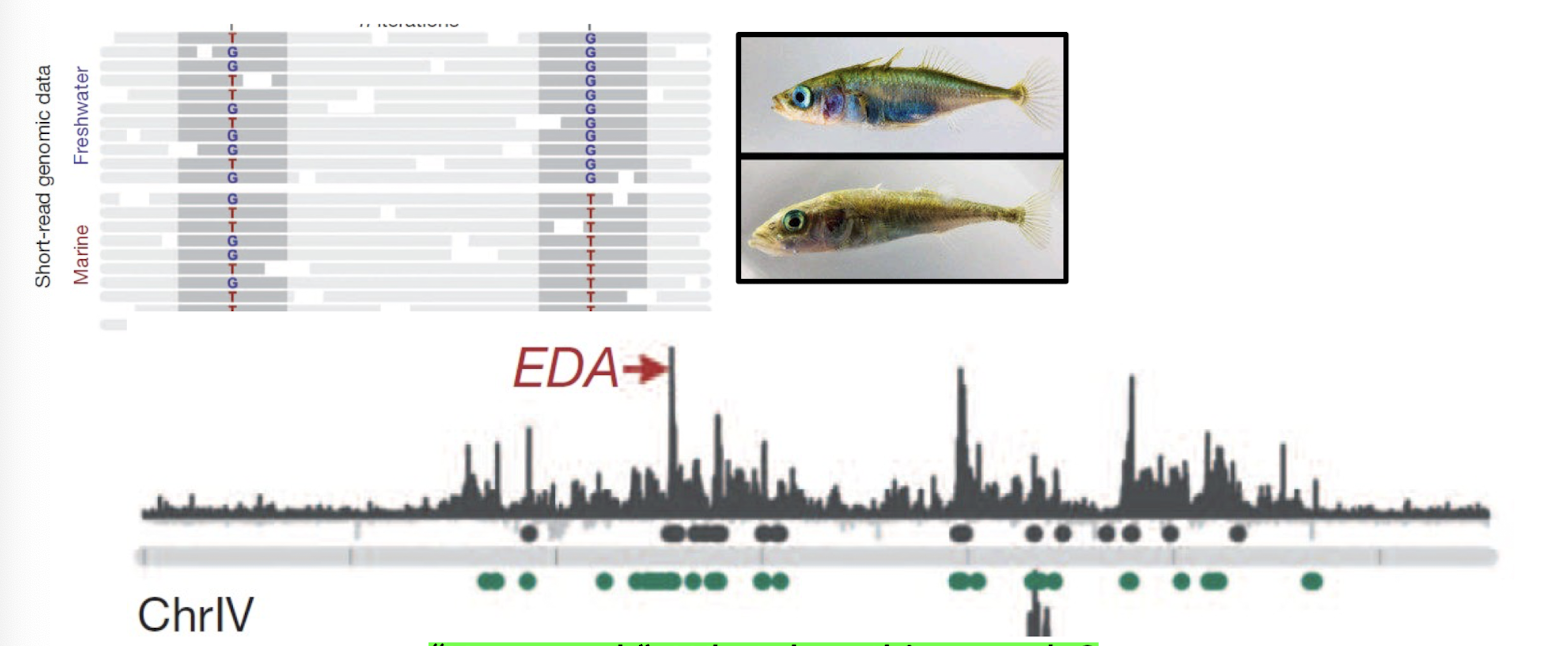
Testing selection with dn/ds or selective sweeps: What gene for the molecular adaptation of Sticklebacks to freshwater environments?
EDA
Short-read genomic data reveals distinct sequence differences between marine and freshwater populations in this gene region
What is a Gene Family?
groups of related genes that evolve via duplication
Members typically share functional and physical (sequence) characteristics
Are members of a gene family identical?
No
unless they have duplicated very recently
How do gene duplications primarily occur?
occur accidentally during DNA replication
Is gene loss always detrimental?
No.
Gene loss can be adaptive, such as in the case of seed shattering in African rice.
What is the difference between "Orthologues" and "Paralogues"?
Orthologues: Genes separated by a Speciation event (e.g., Human Copy 1 and Chimp Copy 1).
Paralogues: Genes separated by a Duplication event (e.g., Human Copy 1 and Human Copy 2)
Why is gene duplication an important consideration for phylogenetics?
If you compare different copies (paralogues) between species instead of the same copy (orthologues)
the resulting tree will reflect the duplication history rather than the true species relationships
2 Gene family size
Small gene families, e.g. human haemoglobins (10 genes)
Large gene families, hundreds of members
What causes gene family size increase or decrease?
Selection
with
– Lineage-specific losses
– Lineage-specific duplications
After Gene Duplication:
What happens if having 2 copies provides a selective disadvantage (or no advantage)?
One copy accumulates mutations and becomes a Pseudogene (gene degradation)
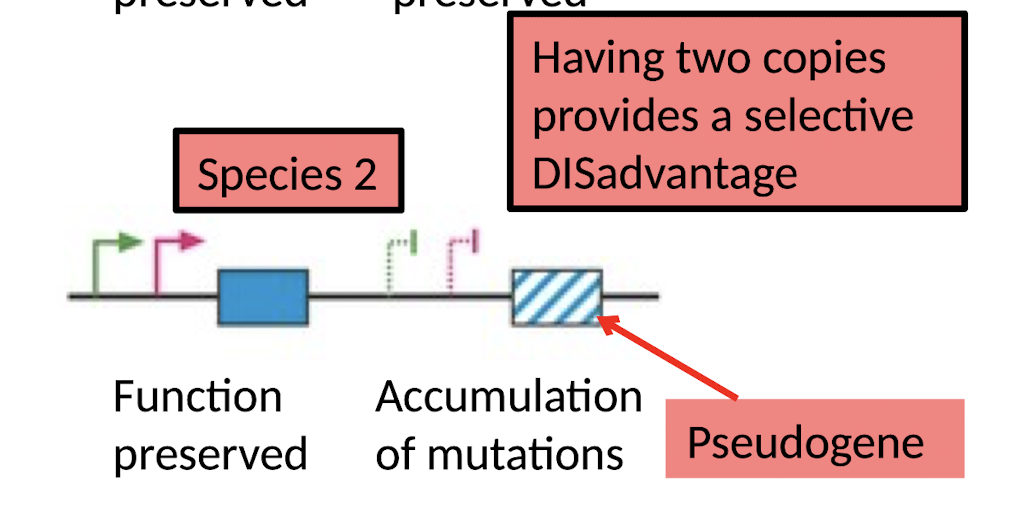
After Gene Duplication:
What happens if having 2 copies provides a selective advantage (e.g., dosage balance)?
Function preserved in both copies
e.g. pheromone-sensing gene in mammals
→ related to neo-functionalisation and sub-neofunctionalisation
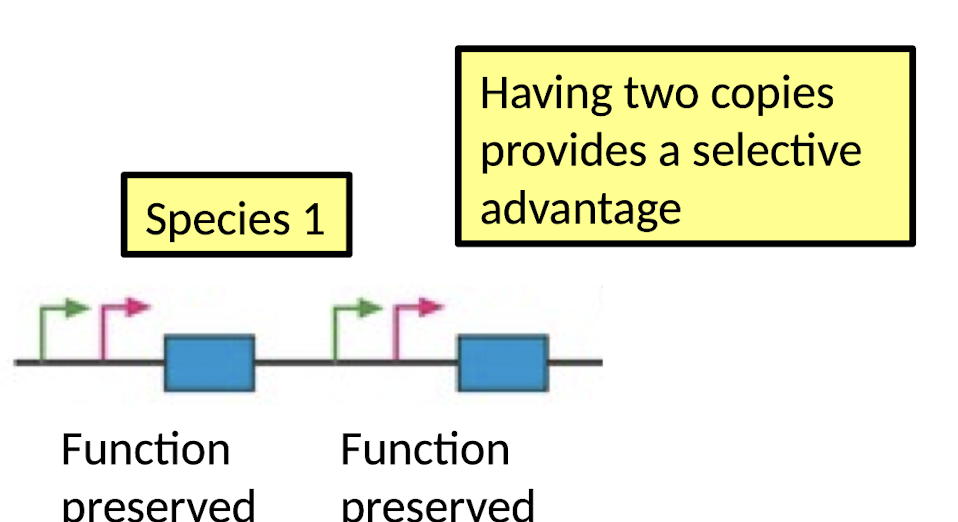
Why is there gene degradation?
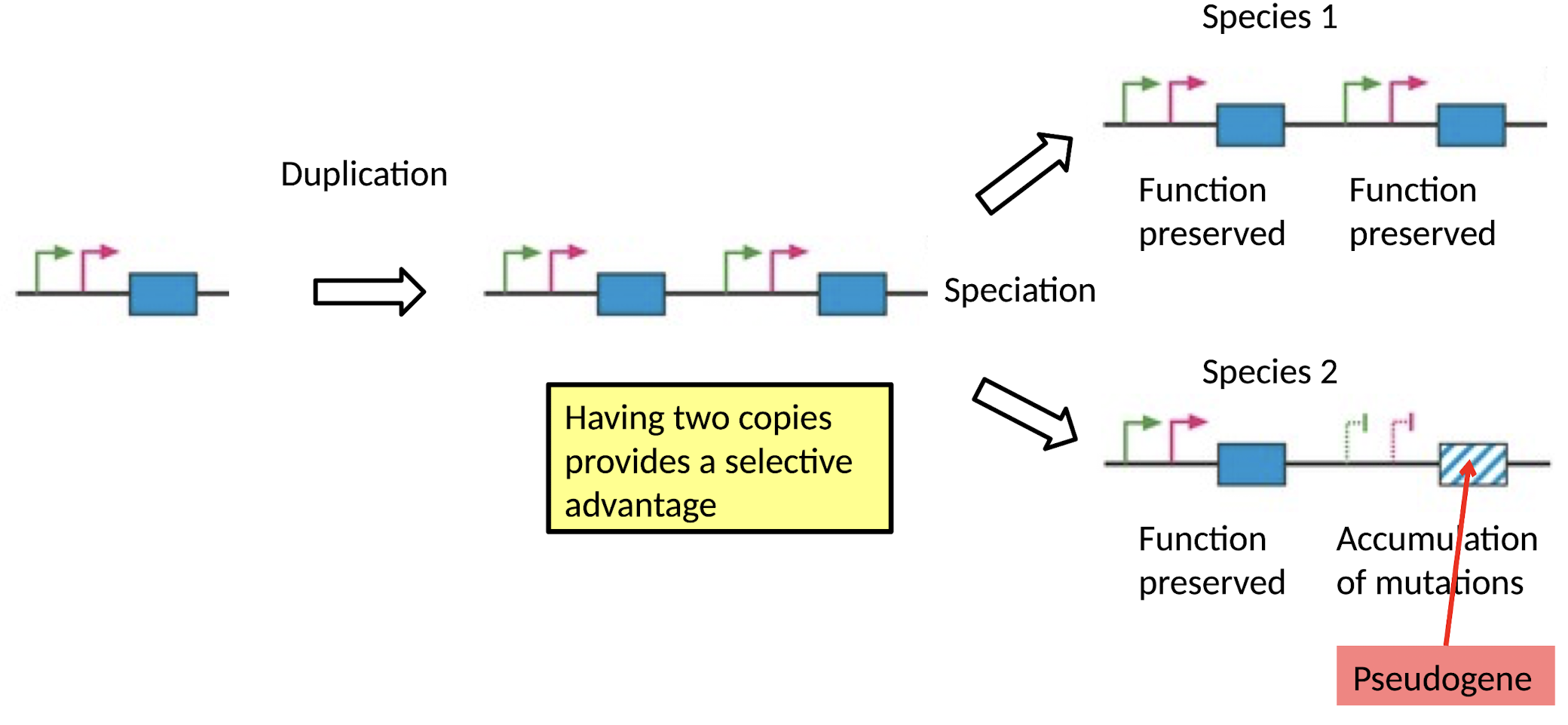
Dosage balance
Natural selection
What is Neofunctionalisation?
when one copy retains the original function, and the duplicated copy evolves a (new) novel function
What example of Neofunctionalisation is given in Antarctic fish?
Ancestral gene (SAS): Synthesizes sialic acids.
New gene (AFPIII): Evolved to function as an Anti-freeze protein that binds to ice crystals.
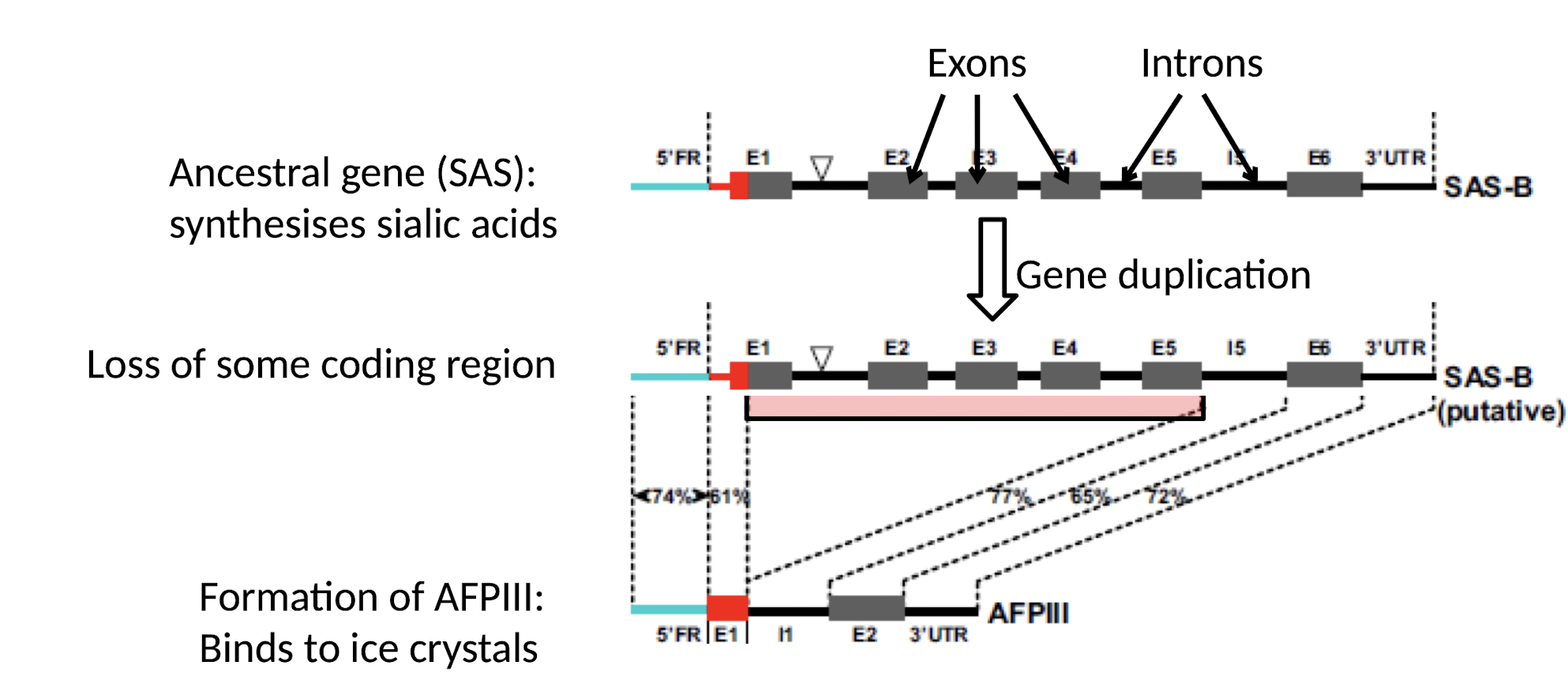
What is Sub-functionalisation?
when original functions of the ancestor gene are partitioned (split) between the duplicated copies
What examples of Sub-functionalisation are provided?
Hemoglobin A in rodents:
Mouse → Diverged in amino acid sequence and changes in O2 binding ability in rat & deermouse
CYCLOIDEA genes in sunflower (Chapman et al (2008,2012):
Many gene duplications → changes in gene expression

How does gene family size change in relation to adaptation?
Gene families can
expand (via duplication)
contract (via loss) depending on selective pressures
What does neo- and sub-functionlisation do to gene family size?
increase in size
What are the specific examples of adaptive gene family expansion mentioned?
Digestive enzymes in fungi:
Fungi adapted to feed on animals (pathogenic) have different gene counts compared to plant-feeders
Neurotoxins: Expanded in scorpions.
Pathogenicity: Expanded in nematode-trapping fungi.
Hypoxia response: Expanded in yaks
What was King and Wilson's (1975) hypothesis regarding Human vs. Chimp evolution?
Regulatory mutations (changes in gene expression) may account for the biological differences between the species
→ as Macromolecules (proteins) are so alike
How does the "Beach vs. Mainland mouse" example illustrate expression evolution?
Phenotype:
Beach mice are light; Mainland mice are dark.
Mechanism: The coding sequence of the Agouti gene is identical.
The difference is caused by sequence differences nearby (regulatory regions) that lead to very high expression in beach mice and low expression in mainland mice
How does the teosinte-branched 1 (tb1) gene differ between Maize and Teosinte?
Coding sequences are the same
BUT expression differs due to the insertion of transposable elements ("Tourist" and "Hopscotch") into the regulatory region of the maize gene
How 2 individuals of the SAME species can differ?
in terms of:
–Nucleotide polymorphisms
–Insertions and deletions
–Gene copy number
–Gene expression
What are the general characteristics of Bacterial Genomes compared to Eukaryotes?
They are compact with very little intergenic sequence (average 116 bp)
They have shared promoters (operons)
They have very low amounts of repetitive DNA (e.g., E. coli has 0.7%, whereas humans have ~50%)
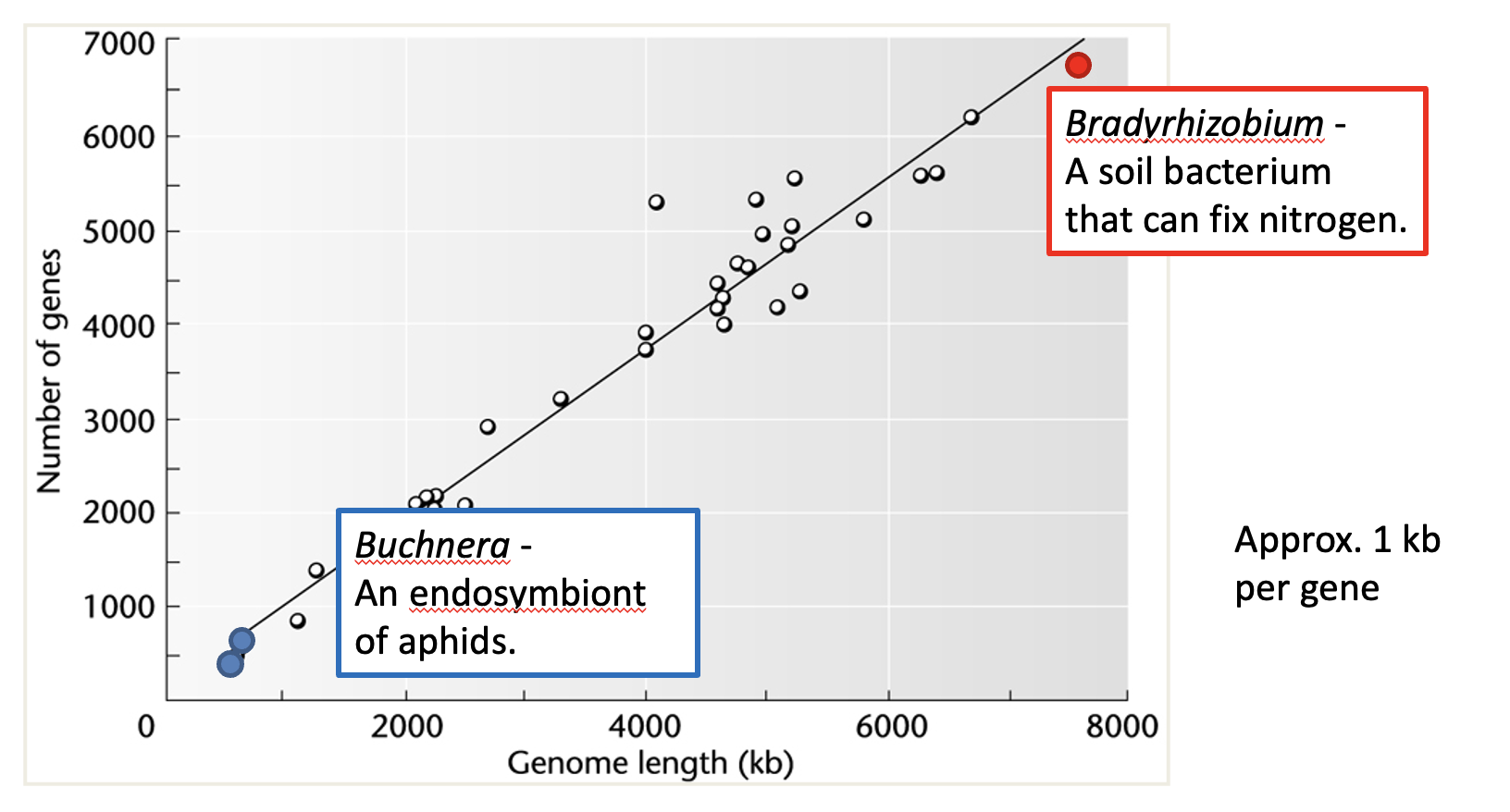
What is the relationship between genome length and gene number in bacteria?
Number of genes is proportional to the genome size
averaging approximately 1 kb per gene
Intraspecific bacteria genome evolution:
How can gene number vary within a single species of bacteria ?
Different strains can have significantly different numbers of genes
Example: E. coli K12 (non-pathogenic) has 4,288 genes, while E. coli O157 (enterohemorrhagic) has 5,060 genes .
Mechanism: A large number of genes may be found in one strain but not others (up to ~1000+ unique genes per strain).
Regulation of bacterial genomes
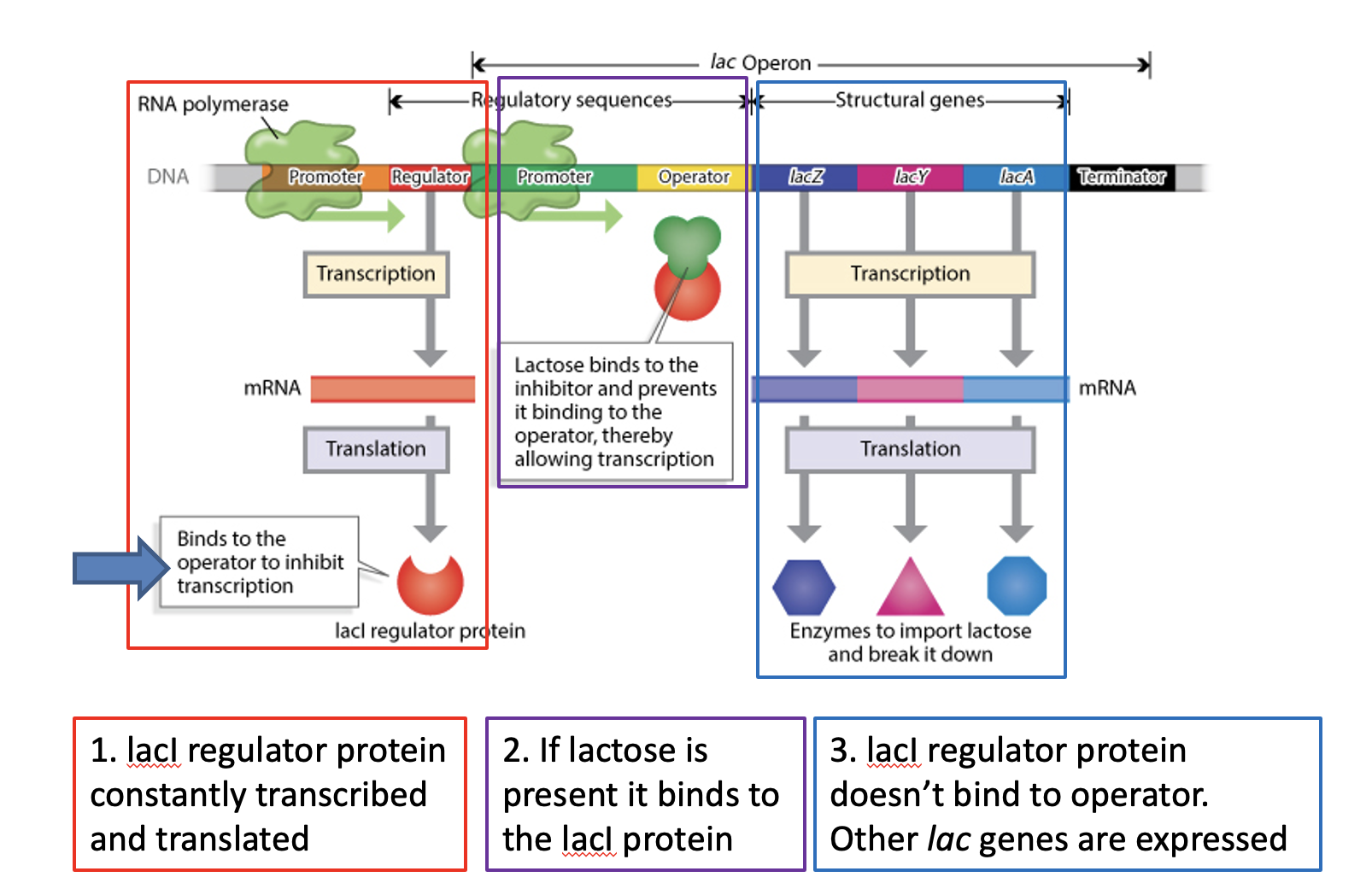
Intraspecific bacteria genome evolution:
What are the two main mechanisms of Horizontal Gene Transfer (HGT) ?
Conjugation: Plasmids (e.g., carrying antibiotic resistance) are transferred between live bacteria .
Transformation: Live bacteria take up DNA released from dead bacteria and incorporate it into their genomes
(more in lecture on HGT)
Intraspecific bacteria genome evolution:
What example is given for bacterial genome size increase and decrease, and what drives it?
Organism: Bradyrhizobium (a nitrogen-fixing soil bacterium).
Genetic Basis: It has a large genome (9,100 kb) containing a 410 kb "symbiosis region". This region contains clusters of genes required for nitrogen fixation (nodulation) to live symbiotically with legumes
Importance: economically important (peas and beans grown on 1.5M km2 farmland per yr)
Organism: Buchnera (an endosymbiont of aphids)
Genetic Basis: It has a very small genome (641 kb, ~1/7th of E. coli).
Lost genes for the biosynthesis of non-essential amino acids because it relies on the host
Many genes for amino acids essential to the aphid
OBLIGATE ENDOSYMBIONT (provide host w what it needs)
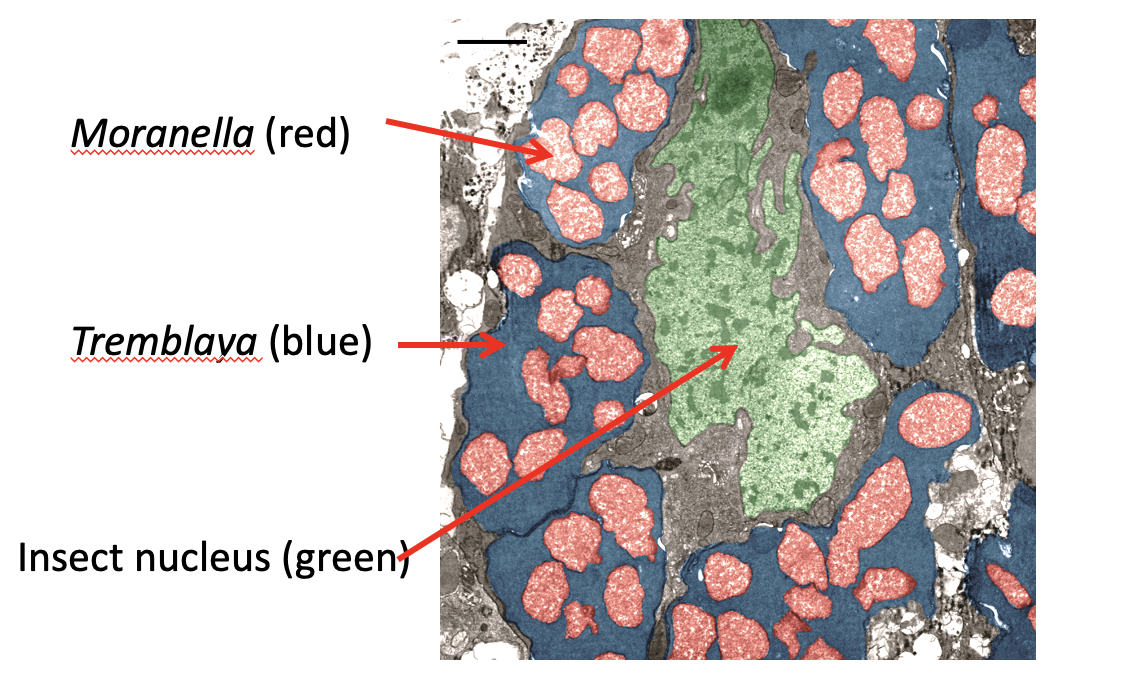
What is the extreme example of genome reduction described as "Bacteria in bacteria"?
"Bacteria in bacteria"
Moranella (538 kb) lives inside Tremblaya (139 kb)
both live inside an insect
Mechanism: They share biochemistry (metabolic pathways like tryptophan and arginine synthesis are split between them), allowing for massive genome reduction

What are the key features of Eukaryotic genomes that contrast with bacteria?
Many Transposable Elements (TEs) and have Repetitive DNA
They possess Introns
Genome size is not proportional to gene number (e.g., yeast vs. humans)