Lecture 4: DNA Replication and Telomere Maintenance
1/27
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
28 Terms
1. Semiconservative model of DNA replication (ON TEST)
Three possible modes of replication hypothesized based on Watson and Crick’s model:
Semiconservative (one strand transfered to next gen)
Conservative (uses the model to make another dna)
Dispersive
Molecuecule broken into pieces and then randoomely inserted into 2 molecules (intertwined)
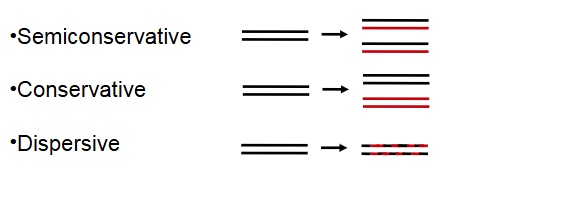
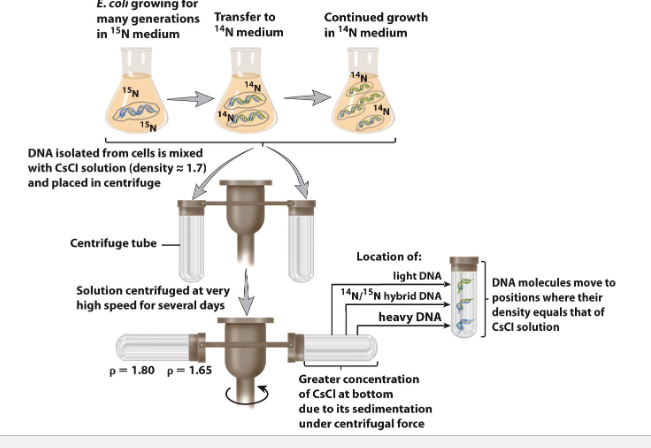
The Meselson-Stahl experiment demonstrated that DNA replication is semiconservative
14N is most abundant form of N
grow ecoli with 14N, as all DNa made of 14N
exp
grow ecoli in the highly isotope N15 media then transfered to 14N media and allow to grow. The add CsCL(density 1.7) then centrifuge to get seperation between light(14N, top) and heavy(15N, bottom) dna
CsCl is used as a resolution to mix them
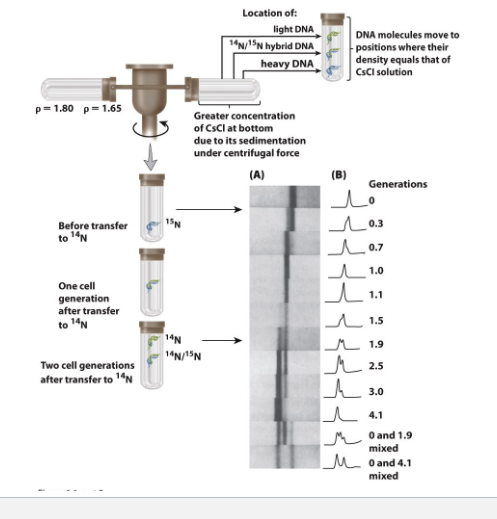
Meselson experiment for semi/comservative (exam)
know band for each and ratio

DNA Replication in Prokaryotes
DNA polymerases are the enzymes that catalyze DNA synthesis from 5′ to 3′
ONLY add nucleotides in 5′ → 3′
CANNOT initiate DNA synthesis de novo.
REQUIRES a primer with free 3’-OH group at end
Deoxynucleoside 5′ triphosphates (dNTPs) added one at a time to the 3′ hydroxyl end of the DNA chain
dNTP add determined by complementary base paring
As phosphodiester bonds form, the two terminal phosphates are lost, making the reaction essentially irreversible.
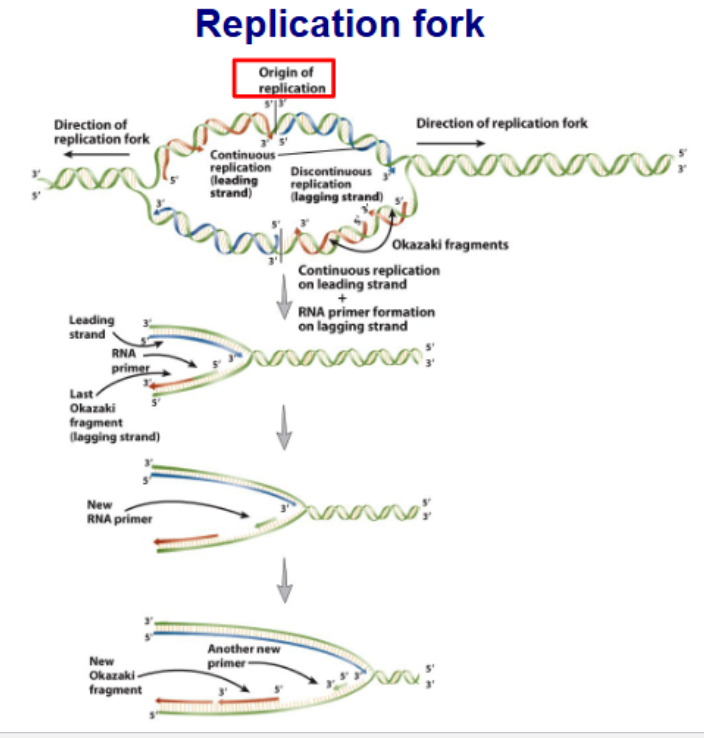
Leading vs Lagging strand
Leading
made continuously in same direction as replication fork’
Lagging Strand
made semi-coontinuously in opp direction as replication form.
DNA made in short segments called Okazaki fragments
→ Nucleotides added to both strands at same time and rate by 2 DNA polymerase
Replication is mediated by replisome
Helicase
unwinds the parental double helix
2 molecules of DNA polymerase 3
Primase (RNA Primer- transcription)
initiates lagging strand Okazaki fragments
2 sliding clamps
attach DNA polymerase to DNA
clamp loader
use ATP to open/close sliding clamps
SSB (single-strand DNA binding proteins
protect DNA from nuclease attack
stabalise single strand DNA template
Topoisomerase
release strains caused by upwinding/winding DNA
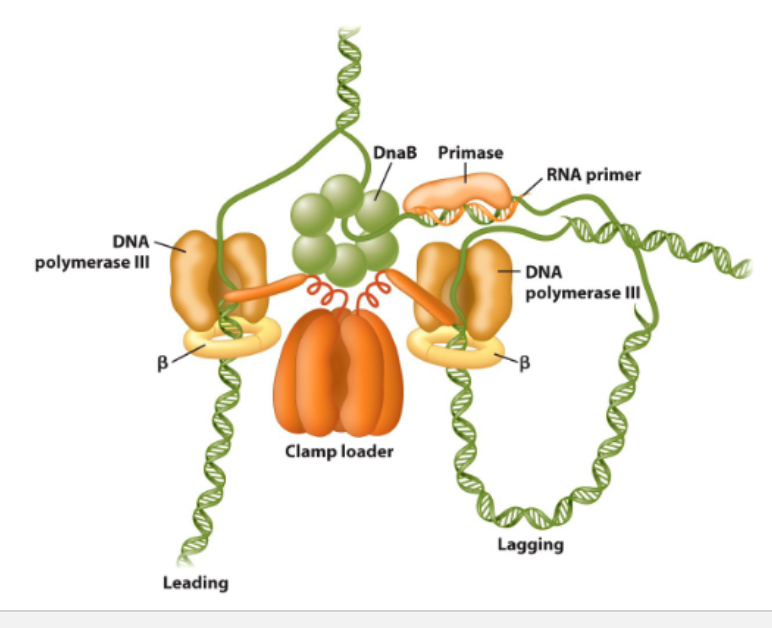
Multi-protein machines mediate bacterial DNA replication
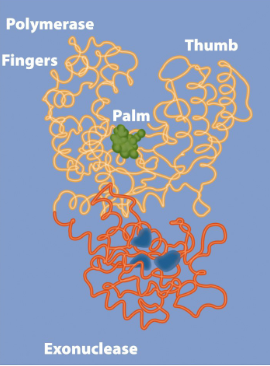
Bacterial DNA polymerases have multiple functions
DNA polymerase I
5′→3′ Polymerase to synthesize DNA (very short)
3′→5′ exonuclease(remove nucleotide) to backtrack (proofreading)
5′→3′ exonuclease to remove primer (DNA repair)
DNA polymerase III
Main replicative polymerase, more processive.
5′→3′ Polymerase to synthesize(ADD) DNA (very short)
3′→5′ exonuclease to backtrack (proofreading)
Initiation of replication
origin
where bidirectional replication fork initiates
some bacteria have a single, well-defined origin
Topoisomerases relax supercoiled DNA
Positive supercoiling ahead of the fork and negative supercoiling in the wake of the fork
DNA topoisomerase
helps relieve torsinal strain that inhibits fork movements
Lagging strand synthesis by the replisome
As replication fork advances the lagging strand polymerase stays with a loop
DNA polymerase 1 remove RNA primers and replace them with complementary dNTPs
DNA ligase catalyzes formation of phosphodiester bond btw adjacent Okazaki fragments.
DNA replication in Eukaryotes
in nucleas, dna organised in nucleosome, and it has linear dna, and need to remve histones, dna is large and needs multiple origins
Potential issues:
1: Nucleosome
2: Linear DNA
3: Multiple origins
Prereplication complex formation and replication licensing
Replication restricted to S phase of cell cycle
Origin selection sep from initiation
prevents overreplication of the genome
RNA priming of leading and lagging strand DNA synthesis
DNA polymerase (pol) “alpha” and its associated primase activity
synthesizes RNA primers in eukaryotes
Polymerase Switching
Def- hand-off of DNA template from one polymerase to another
Leading strand: switch from DNA polymerase “alpha” to pol “epsilon”
* Lagging strand: switch from pol “alpha” to pol “Delta”
Proofreading
Replicative polymerases are high fidelity but not perfect: 10-4 to 10-5 errors per base pair.
Proofreading using 3' to 5' exonuclease activity reduces the error rate to 10-7 to 10-8 errors per base pair
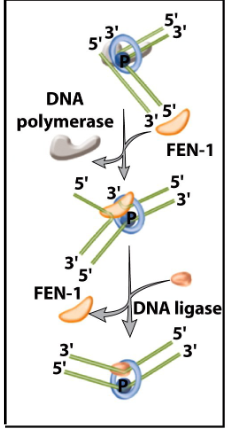
Removal of primers
RNA primer removed by FEN-1 and/or Rnase H
Topoisomerase untangles the newly synthesized DNA
in eukaryotes: rep continues until 1 form meets adjacent replicon fork
progency DNA molecules remain intertwined
Topoisomerase 2 req to resolve the two separate progeny genomes
Telomere maintenance
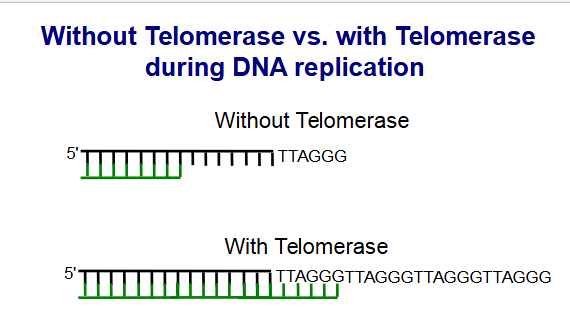
The end replication problem
When final primer removed, a 8-12 nucleotide region is left Un-replicated. which predicts that chromosomes get shorter with each replication round
Telomers
Eukaryote chromosomes end with tandem repeats of a simple G-rish sequence
Humans: TTAGGGG
Tetrahymena: TTGGGG
seal ends of chromosomes
creates stability by keeping chromosomes from ligating together
Carol Greider and Elizabeth Blackburn discovered Telomere
studied etrahymena thermophila, a single-celled eukaryote with over 40,000 telomeres
DISCOVERED the enzyme TELOMERASE
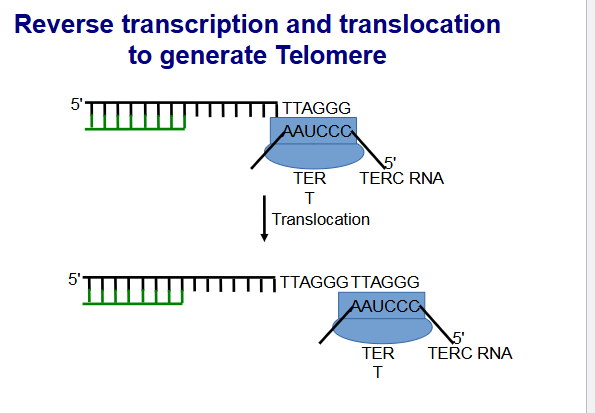
Maintenance of telomeres by telomerase
Telomerase elongates the 3′ end of the template for the
lagging strand (G-rich overhang).Telomerase is a ribonucleoprotein (RNP) complex with
Telomerase reverse transcriptase (TERT) activity.Contains an Telomerase RNA component (TERC) that
provides the template for telomere repeat synthesis.epeated translocation and elongation steps results in
chromosome ends with an array of tandem repeats.
with the telomerae the copy gets made adn then added to stop cell death
Regulation of telomerase activity
telomera length involves accessibility of telomeres to telomerase
length control factors include
proteins POT1, TRF1, TRF2
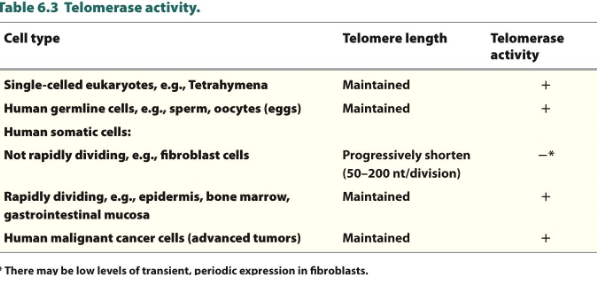
Telomerase, aging, and cancer
telomerase Has HOUSEKEEPING function- in most unicellular organisms
Progressive shortening of telomeres
in most human somatic cells not enough telomerase is expressed to maintain length
High levels of telomerase activity in ovaries, testes, rapidly dividing somatic cells, and cancer cells.
Telomere shortening: a molecular clock for aging?
Telomerase- target of anti-aging/ or cancer therapy
in cancer cells
telomerase has been reactivated
Telomerase and aging: the Hayflick limit
The Hayflick limit is the point at which cultured cells stop dividing and enter an irreversible state of cellular aging (senescence).
Proposed to be a consequence of telomere shortening