05 - RNA Structure & Transcription
1/59
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
60 Terms
RNA
- Is a single strand - Has ribose instead of deoxyribose - Has uracil instead of thymine
DNA & RNA both have…
- A sugar/phosphate backbone and nitrogenous bases
- 5’-3’ directionality within a strand
- Complementary base-pairing
Structure of RNA
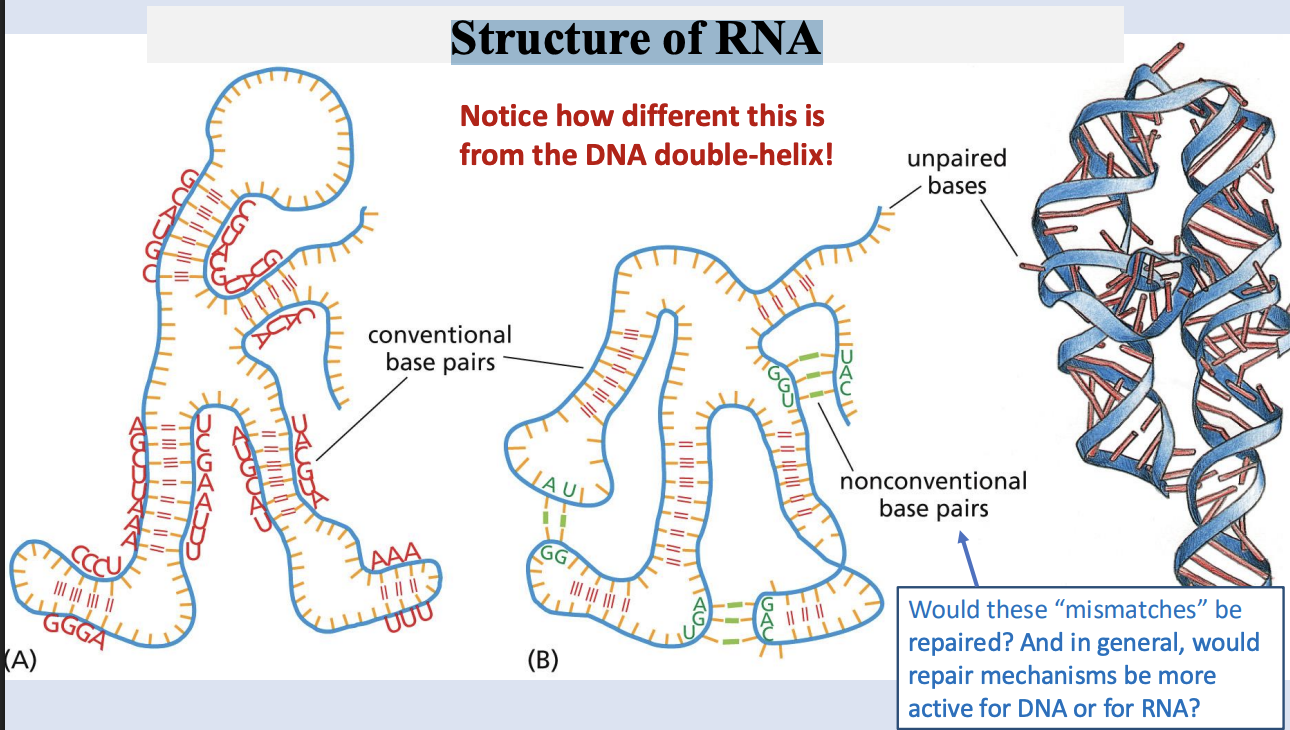
primary structure of rna
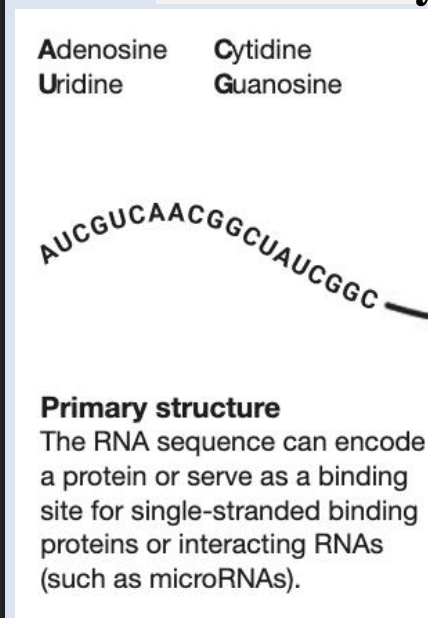
secondary structure of rna
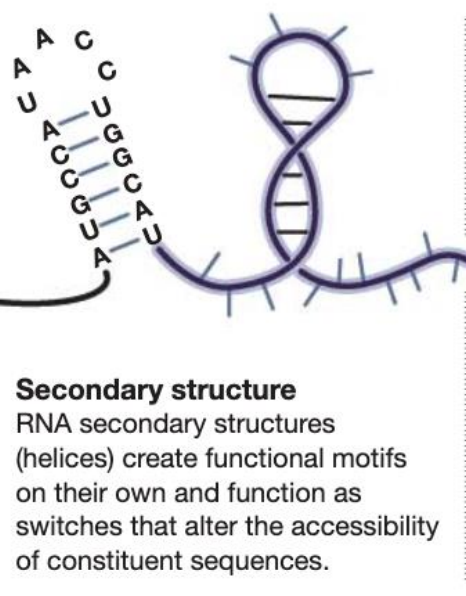
tertiary structure of rna
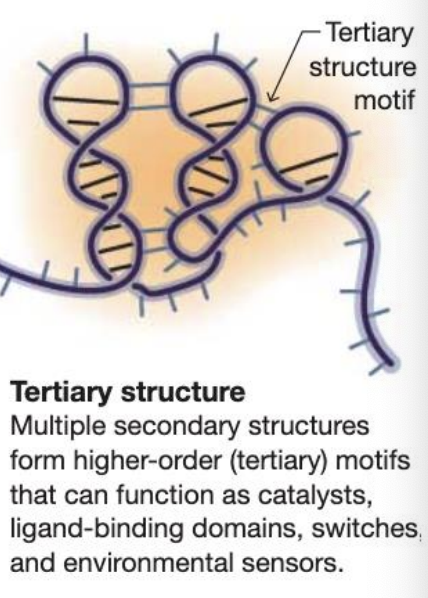
Messenger RNA (mRNA)
The intermediate between DNA and protein – used as a template for protein synthesis by ribosomes in cytoplasm
Ribosomal RNA (rRNA)
Provide structural support in the ribosome and catalyzes the chemical reaction in which amino acids are covalently linked to each other
Transfer RNA (tRNA)
Required for translation of mRNA information into polypeptide sequence i.e. convert the language of nucleotides to the language of amino acids.
rrna trna and mrna lives length
rRNAs and tRNAs are long lived and have complex secondary and tertiary structures that play a role in their functions. mRNA have shorter half-lives
Isn’t it more work for the cell if the mRNA degrades and needs to be replaced? Why wouldn’t you want the RNA that codes for protein to be the most stable?
to contrtol and regulate the amount of protein being produced
more stable rna produces more protein
Transcription: production, how it is cairried out
produces RNA complementary to one strand of DNA
Carried out by enzymes called DNA-dependent RNA polymerases
Go from information storage (DNA) to information use (RNA and proteins) in the cell
Direction of transcription is determined by
the orientation of the promoter sequence in the DNA at the beginning of each gene
What do you mean the “orientation” of the promoter?
The “orientation” of the promoter refers to the direction in which the promoter drives transcription on the DNA strand. Promoters are not symmetrical, so they can only initiate transcription in one direction.
Flip the promoter = you transcribe the wrong strand in the wrong direction, possibly producing a nonsense RNA or no functional transcript at all.
Promoter region in prokaryotes
-10 and -35 element
Three main stages of transcription
Initiation
Elongation
Termination
Initiation
RNA polymerase binds to a specific sequence of nucleotides (promoter sequence). This requires the help of transcription factors (proteins)
Ribonucleoside triphosphates (rNTPs) are the building blocks instead of deoxyribonucleoside triphosphates (dNTPs) from DNA synthesis
Elongation
Open up DNA strands temporarily allowing synthesis of complementary RNA. Ribonucleotides are linked together by phosphodiester bonds (5’ to 3’ like DNA synthesis!)
Termination
In prokaryotes, RNA polymerase reaches a specific signal sequence on the DNA that causes an extended pause in synthesis and release of the RNA.
RNA polymerase with rna and dna
• DNA makes a sharp turn within the active site of the enzyme
• DNA and RNA exit out of separate channels
• Note that only a small portion of DNA is single-stranded at a time
Sigma Factor
increases the RNA pol’s affinity for promoter sequences. Different σ factors can lead to expression of different genes
RNA Polymerase Holoenzyme
RNA Polymerase (5 subunits) + sigma factor (σ) is called the RNA Polymerase Holoenzyme
Transcription termination in Prokaryotes
• Termination sequences stop transcription.
• Can be “intrinsic” when stem-loop hairpins cause RNA pol to pause
• Can be protein-mediated by providing a binding sequence for a protein.
(Ex. A protein called rho factor (ρ) in bacteria)
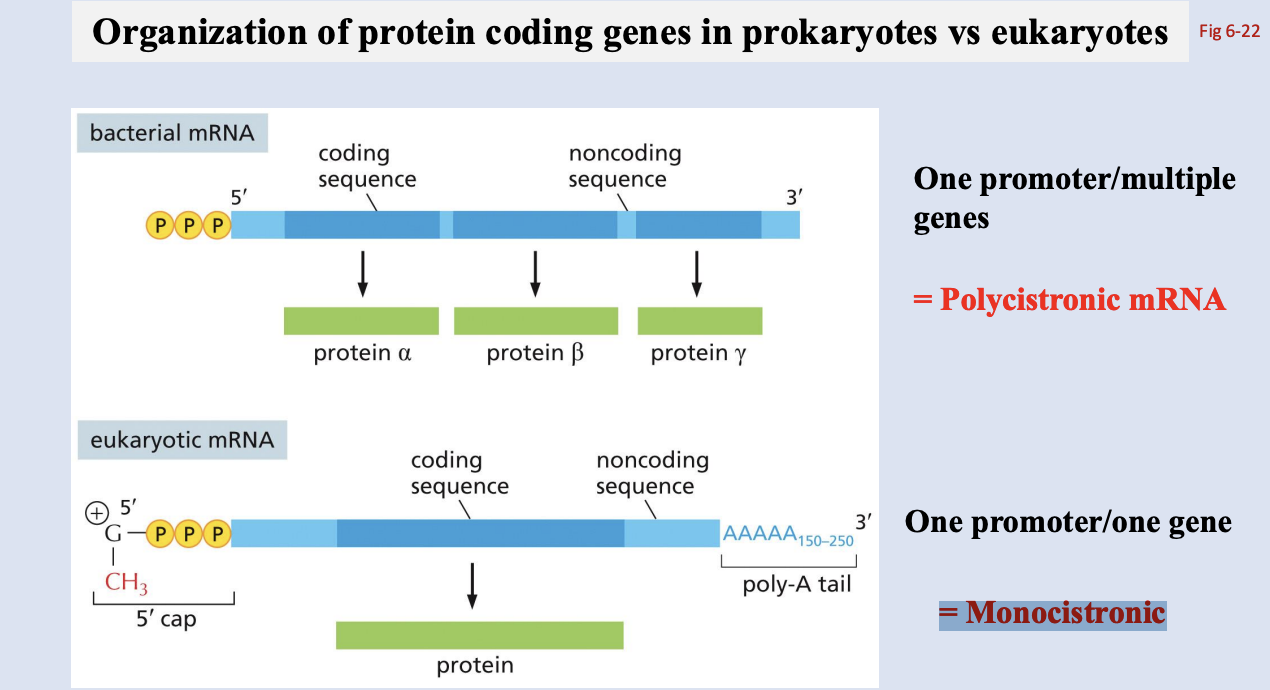
Organization of protein coding genes in prokaryotes vs eukaryotes
bacterial mRNA: One promoter/multiple genes = Polycistronic mRNA
Transcription and translation coupled
eukaryotic mRNA: One promoter/one gene = Monocistronic
Transcription and translation uncoupled
What benefits could there be to using polycistronic mRNA in prokaryotes?
saves space and easier coordination (efficiency in regulation)
Transcription in Eukaryotes vs Prokaryotes: rna processing
Eukaryotic RNA requires processing in the nucleus (5’ cap, splicing, 3’ poly A tail)
Export: RNA is made in the nucleus, but mRNA will be translated in the cytoplasm
Alternative splicing
allows 1 gene to be used for production of multiple different sequences of proteins (splice isoforms)
RNA sequences + proteins regulate this process
What are potential advantages to alternative splicing?
many sets of messsages and protein products can get produced from a limited number of genes (efficiency and smaller genome size)
Transcription in Eukaryotes
Three major types of RNAs (rRNA, mRNA, tRNA) as well as microRNAs are derived from precursors that are larger than the final RNA product (not true in prokaryotes)
microRNAs derived from precursors process
Transcriptional unit encoded in the DNA includes promoter, coding sequence and sequences for termination of transcription
A primary transcript (or pre-RNA or precursor-mRNA) is the initial RNA molecule synthesized from the template DNA
RNA Polymerase II synthesizes all eukaryotic mRNA precursors (premRNA)
RNA Polymerases in Eukaryotes
rna polymerase I
rna polymerase II
rna polymerase III
general transcription factors, (GTFs)
Transcription in eukaryotes requires general transcription factors, (GTFs)
Additionally, the pattern / amount of transcription is regulated by presence of other transcription factors (and associated proteins) that bind DNA regulatory sequences (i.e. enhancers)
Consensus sequences near eukaryotic RNA pol II start points
These are short conserved DNA sequences found upstream or around the transcription start site (TSS).
They help recruit transcription machinery like general transcription factors (GTFs) and RNA Pol II.
Transcription in Eukaryotes: TATA box
promoter sequence is -24 to -32 from the transcription start site. (- 30 in humans). Sequence is either identical or similar to 5’-TATAAA-3’
site of assembly of preinitiation complex (PIC) that contains GTFs and RNA polymerase
assembly of preinitiation complex (PIC)
1. TFIID recognizes and binds the TATA box through its subunit TBP (TATA binding protein), which binds and bends the DNA
2. Enables other GTFs (TFIIA and TFIIB) to also bind
3. RNA polymerase is recruited to the PIC along with TFIIF, followed by TFIIE and TFIIH
TFIIH
(kinase and helicase) uses ATP hydrolysis to open the DNA double helix at the transcription start point, to expose single-stranded template
TFIIH phosphorylates the RNA polymerase II tail (called the C-terminal domain, CTD), which signals that RNA Pol II can begin elongation.
Other GTFs are released once elongation begins, but TFIID usually stays bound at the promoter to start another PIC
Transcription through nucleosomes
Three transcription elongation factors (Spt4, Spt5, and Elf1) help the polymerase transcribe through nucleosomes.
Also aided by ATP-dependent chromatin remodeling complexes and histone chaperones (that can partially disassemble nucleosomes)
what can transcription elongation factors do
• wedge DNA away from the histone core;
• destabilize histone–DNA interactions;
Anatomy of eukaryotic mRNA
The processed mRNA contains a “coding sequence” of nucleotides that will direct the order of amino acids in the encoded protein. Splicing has removed the _____ RNA
Untranslated regions (5’ UTR or 3’ UTR) are RNA sequences that do not encode protein but play important regulatory roles
5’ end: methylated guanosine cap
3’ end: 50-250 adenosines → Poly(A) tail
During transcription, proteins involved in RNA processing are carried on the RNA Pol II CTD and transferred to the nascent RNA at the appropriate time What are the 3 types of processing that need to occur?
splicing of introns, poly a tail and methyl cap
Processing can begin as soon as the sequences to be processed have been transcribed.
Which type of processing will happen first? Which is last
5’ cap, intron spile and last poly tail
RNA synthesis and processing
Capping proteins are the first to bind the CTD.
RNA molecule is efficiently capped as soon as its 5′ end emerges from the RNA polymerase.
Which CTD sites are phosphorylated switches as RNA pol II continues, which recruits proteins involved in splicing and then 3′-end processing proteins
methylguanosine capping is initiated when
immediately after synthesis of 5’ end of RNA
importance of Addition of 5’ cap
• Cap prevents 5’ end from being digested by exonucleases
• Plays a role is transport out of nucleus and initiation of mRNA translation to protein
• Helps cells to distinguish completed mRNA from other types of RNA molecules
why is it important for cell to distinguish completed mRNA from other types of RNA molecules
to avoid production of unessary protein and spend our translation efforts on our actual mrnas
Capping enzyme
two active sites for two functions → RNA triphosphatase and guanylyl transferase
Addition of 5’ cap steps
Step 1: RNA Triphosphatase removes the terminal phosphate group, leaving a disphosphate
Step 2: Guanyl transferase adds a GMP in an inverted orientation (5’ end of guanosine faces 5’ end of RNA), which forms 5’-5’ triphosphate bridge
Step 3: The guanine base is then methylated by RNA methyltransferase. • The ribose to which the GMP was attached is also methylated.
when is Addition of 3’ poly(A) tail intiated
Addition of a string of A nucleotides occurs at the 3’end of the mRNA → polyadenylation
importance of Addition of 3’ poly(A) tail
This sequence acts as a recognition binding site for assembly of the protein complex that carries out polyadenylation
Protects from digestion by exonucleases
Can be used to isolate mRNA from total RNA using oligo dT column (TTTTTTTT… on column base pairs with tail) (significant tool in molecular biology)
Addition of 3’ poly(A) tail process
CstF & CPSF are initially attached to RNA Pol CTD but then get transferred to the RNA
Initiates cleavage to create a new 3’ end
Poly(A) polymerase can then add adenosines to the new 3’ end, without the need for a DNA template (rare for polymerases!)
Poly-A-binding proteins coat the tail
Each gene can be used to make many copies of RNA and each mRNA can be used to make many copies of protein (t/f)
t
RNA molecules are single-stranded and lack complementary base-pairing (t/f)
f
There are different types of RNA (t/f)
t
Repair mechanisms ensure that RNA does not contain mismatched base pair (t/f)
f
RNA synthesis for all genes in a genome uses the same strand of the double helix as a template (t/f)
f
E. coli transcription terminators contain a -10 and -35 element (t/f)
f
Which of the following are true about making mRNA in eukaryotes?
A) mRNA is transcribed by RNA polymerase II
B) Translation into protein often begins before synthesis of the RNA transcript is complete
C) Transcription terminates when it reaches the TTTTTT… template sequence that encodes the 3’ poly(A)tail
D) 5’ capping occurs while transcription of the rest of the pre-mRNA is still ongoing
E) The RNA pol II CTD is required for forming the pre-initiation complex, but then is cleaved to allow elongation to begin
a and d
Both splice isoforms and an alleles can result in different version of a protein, so what is the difference between them?
Splice isoforms arise from the same gene via alternative RNA splicing, while alleles are different DNA versions of a gene inherited from parents.
If the sense strand of a gene sequence is 5’ ACGTA-3’, then what is the RNA sequence produced from that portion of the gene?
The RNA sequence will be 5’-ACGUA-3’, identical to the sense strand except with U instead of T.
What is a consensus sequence? Why is it a “consensus” instead of just a single, best sequence?
A consensus sequence is a generalized sequence that represents the most common nucleotides at each position among many similar sequences, reflecting variability rather than a single "best" version.