Genetics - Exam 3
1/224
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
225 Terms
ribozymes
catalytic RNA molecules that can cut out parts of their own sequences, connect some RNA molecules together, replicate others, and catalyze the formation of peptide bonds between amino acids
The discovery of ribozymes complements other evidence suggesting that the original genetic material was RNA.
Ribosomal RNA (rRNA) and ribosomal protein subunits
make up the ribosome, the site of protein assembly
Messenger RNA (mRNA)
carries the coding instructions for a polypeptide chain from DNA to a ribosome.
After attaching to the ribosome, an mRNA molecule specifies the sequence of the amino acids in a polypeptide chain and provides a template for the joining of those amino acids.
pre-messenger RNAs (pre-mRNAs)
the immediate products of transcription in eukaryotic cells.
Pre-mRNAs are modified extensively before becoming mRNA and exiting the nucleus for translation into protein.
Bacterial cells do not possess pre-mRNA; in these cells, transcription takes place concurrently with translation.
Transfer RNA (tRNA)
serves as the link between the coding sequence of nucleotides in an mRNA molecule and the amino acid sequence of a polypeptide chain.
Each tRNA attaches to one particular type of amino acid and helps incorporate that amino acid into a polypeptide chain
small nuclear ribonucleoproteins (snRNPs, aka “snurps”)
combo of small nuclear RNAs (snRNAs) and small protein subunits
Some snRNAs participate in the processing of RNA, converting pre-mRNA into mRNA.
small nucleolar RNAs (snoRNAs)
take part in the processing of rRNA
microRNAs (miRNAs) and small interfering RNAs (siRNAs)
carry out RNA interference (RNAi), a process in which these small RNA molecules help trigger the degradation of mRNA or inhibit its translation into protein.
piwi-interacting RNAs (piRNAs; named after Piwi proteins, with which they interact)
Found in mammalian testes, these RNA molecules are similar to miRNAs and siRNAs; they have a role in suppressing the expression of transposable elements in reproductive cells.
Long noncoding RNAs (lncRNAs)
are relatively long RNA molecules found in eukaryotes that do not code for proteins.
They provide a variety of functions, including regulation of gene expression
CRISPR RNAs (crRNAs)
RNA interference-like system
assist in the destruction of foreign DNA molecules
Like replication, transcription requires three major components:
A DNA template
The raw materials (ribonucleotide triphosphates) needed to build a new RNA molecule
The transcription apparatus, consisting of the proteins necessary for catalyzing the synthesis of RNA
template strand
nucleotide strand used for transcription
nontemplate strand
not oridnarly transcribed
transcription unit
stretch of DNA that encodes an RNA molecule and the sequences necessary for its transcription.
whats included in a transcription unit?
a promoter, an RNA-coding region, and a terminator
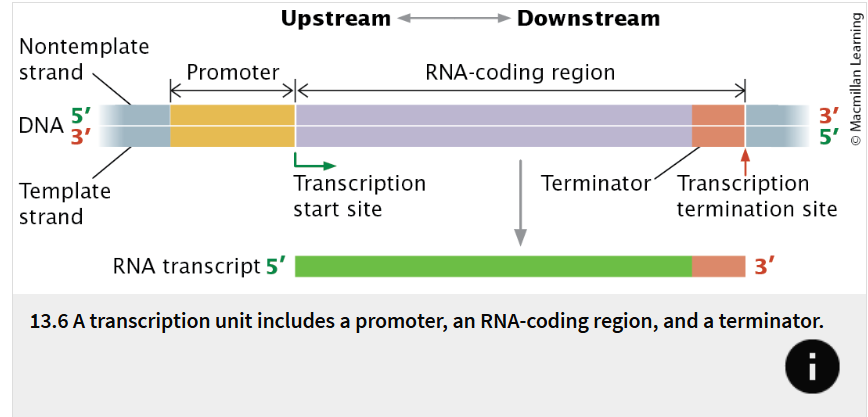
promoter
a DNA sequence that the transcription apparatus recognizes and binds.
The promoter indicates which of the two DNA strands is to be read as the template and the direction of transcription.
It also determines the transcription start site, the first nucleotide that will be transcribed into RNA.
In many transcription units, the promoter is located next to the transcription start site but is not itself transcribed.
RNA-coding region
a sequence of DNA nucleotides that is copied into an RNA molecule.
terminator
a sequence of nucleotides that signals where transcription is to end.
Terminators are usually part of the RNA-coding sequence
transcription stops only after the terminator has been copied into RNA.
ribonucleoside triphosphates (rNTPs)
what RNA is synthesized from
consist of a ribose sugar and a base (a nucleoside) attached to three phosphate groups
overall chemical reaction for the addition of each nucleotide during RNA synthesis
where PPi represents pyrophosphate.
Nucleotides are always added to the 3′ end of the RNA molecule, and the direction of transcription is therefore 5′→3′ , the same as the direction of DNA synthesis during replication.
Thus, the newly synthesized RNA is complementary and antiparallel to the template strand. Unlike DNA synthesis, RNA synthesis does not require a primer.
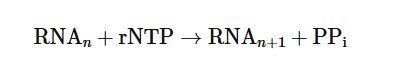
RNA polymerase
Enzyme that synthesizes RNA from a DNA template during transcription.
Bacterial rna polymerase
typically possess only one type of RNA polymerase, which catalyzes the synthesis of all classes of bacterial RNA: mRNA, tRNA, and rRNA.
Bacterial RNA polymerase is a large multimeric enzyme (meaning that it consists of several polypeptide chains).
core enzyme
five subunits at the heart of most bacterial RNA polymerase
two copies of a subunit called alpha (α) and single copies of subunits beta (β), beta prime (β′), and omega (ω)
The ω subunit is not essential for transcription, but it helps stabilize the enzyme.
The core enzyme catalyzes the elongation of the RNA molecule by the addition of RNA nucleotides. Other functional subunits join and leave the core enzyme at particular stages of the transcription process.
sigma (σ) factor
controls the binding of RNA polymerase to the promoter.
Without sigma, RNA polymerase initiates transcription at a random point along the DNA
after the sigma factor has associated with the core enzyme, RNA polymerase binds stably only to the promoter and initiates transcription at the proper start site.
Sigma is required only for promoter binding and initiation; after a few RNA nucleotides have been joined together, sigma usually detaches from the core enzyme.
Many bacteria have multiple types of sigma factors; each type initiates the binding of RNA polymerase to a particular set of promoters.
holoenzyme
forms after the sigma factor has associated with the core enzyme
binds to the -35 and -10 consensus sequences
Although it binds only the nucleotides of the consensus sequences, the enzyme extends from −50 to +20 when bound to the promoter.
The holoenzyme initially binds weakly to the promoter but then undergoes a change in structure that allows it to bind more tightly and unwind the double-stranded DNA
Unwinding begins within the −10 consensus sequence and extends downstream for about 14 nucleotides, including the start site (from nucleotides −12 to +2).
RNA polymerase I
transcribes rRNA in eukaryotes
RNA polymerase II
transcribes pre-mRNAs, snoRNAs, some miRNAs, and some snRNAs in eukaryotes
RNA polymerase III
transcribes other small RNA molecules—specifically tRNAs, small rRNAs, some miRNAs, and some snRNAs
RNA polymerase IV
transcribes siRNAs that silence transposons in plants
RNA polymerase V
transcribes siRNAs that play a role in DNA methylation and chromatin structure in plants
α-Amanitin
a potent inhibitor of RNA polymerase II
it binds to RNA polymerase and jams the moving parts of the enzyme, interfering with its ability to move along the DNA template.
In the presence of α-amanitin, RNA synthesis slows from its normal rate of several thousand nucleotides per minute to just a few nucleotides per minute.
The results are catastrophic, leading to cell death.
The liver, where the toxin accumulates, is irreparably damaged and stops functioning. In severe cases, it causes death.
Rifamycins
a group of antibiotics that kill bacterial cells by inhibiting RNA polymerase.
These antibiotics are widely used to treat tuberculosis
The structures of bacterial and eukaryotic RNA polymerases are sufficiently different that rifamycins can inhibit bacterial RNA polymerases without interfering with eukaryotic RNA polymerases.
Research has demonstrated that several rifamycins work by binding to the part of the bacterial RNA polymerase that clamps onto DNA and then jamming the RNA polymerase, thus preventing RNA polymerase from interacting with the promoter on the DNA.
three stages of transcription:
Initiation, in which the transcription apparatus assembles on the promoter and begins the synthesis of RNA
Elongation, in which DNA is threaded through RNA polymerase and the polymerase unwinds the DNA and adds new nucleotides, one at a time, to the 3′ end of the growing RNA strand
Termination, the recognition of the end of the transcription unit and the separation of the RNA molecule from the DNA template
Initiation
(1) promoter recognition, (2) formation of a transcription bubble, (3) creation of the first bonds between rNTPs, and (4) escape of the transcription apparatus from the promoter.
consensus sequence
the set of the most commonly encountered nucleotides among sequences that possess considerable similarity, or consensus
-10 consensus sequence
most common consensus sequnce found in almost all bacterial promoters
centered 10 bp upstream of the start site
TATAAT
Remember that TATAAT is just the consensus sequence—representing the most commonly encountered nucleotides at each of these sites
In most prokaryotic promoters, the actual sequence is not TATAAT.

-35 consensus sequence
another consensus sequnce common to most bacterial promoters
TTGACA
about 35 nucleotides upstream of start site
down mutations
base substitutions within the −10 and −35 consensus sequences
reduce the rate o ftranscription
up mutation
Occasionally, a particular change in a consensus sequence increases the rate of transcription
upstream element
contains a number of A–T pairs and is found at about −40 to −60.
A number of proteins may bind to sequences in and near the promoter; some stimulate the rate of transcription and others repress it.
abortive initiation
RNA polymerase repeatedly generates and releases short transcripts, from 2 to 6 nucleotides in length, while still bound to the promoter
occurs in both prokaryotes and eukaryotes.
After several abortive initiation attempts, the polymerase synthesizes an RNA molecule from 9 to 12 nucleotides in length, which allows the polymerase to transition to the elongation stage.
backtracking
disengages the 3’OH group of RNA molecule from active site of RNA polymerase and temprorarily halts further RNA synthesis
important in transcriptional proofreading
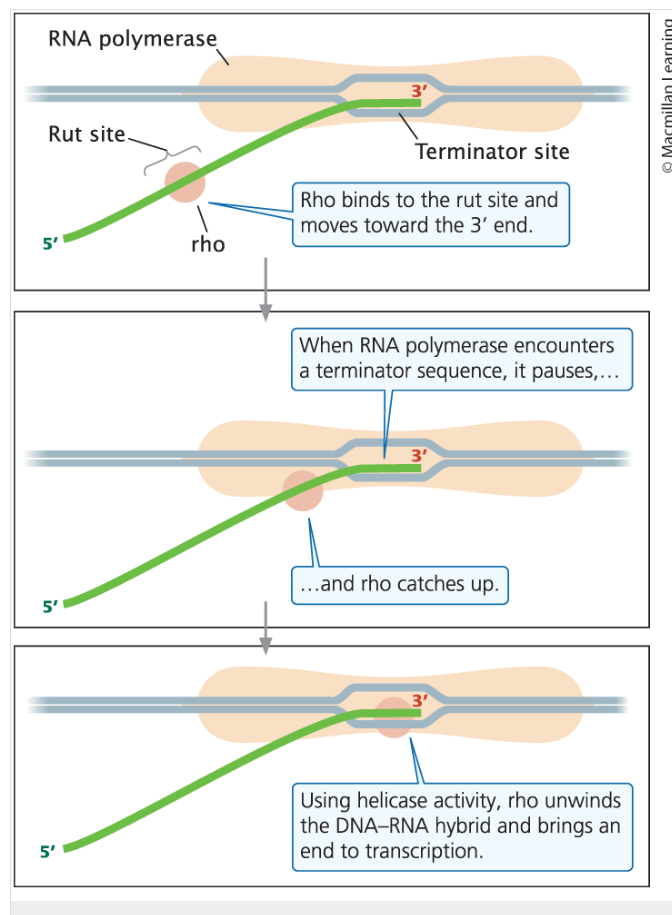
Rho-dependent terminators
able to cause the termination of transcription only in the presence of an ancillary protein called the rho factor (ρ)
Rho-dependent terminators have two features.
The first is the DNA sequence of the terminator itself; this sequence causes the RNA polymerase to pause.
The second feature is a DNA sequence upstream of the terminator that encodes a stretch of RNA that is usually rich in cytosine nucleotides and devoid of any secondary structures.
this RNA sequence is called the rho utilization (rut) site
Once rho binds to the RNA, it moves toward its 3′ end, following the RNA polymerase
When RNA polymerase encounters the terminator, it pauses, allowing rho to catch up.
The rho factor has helicase activity, which it uses to unwind the DNA–RNA hybrid in the transcription bubble, bringing transcription to an end.
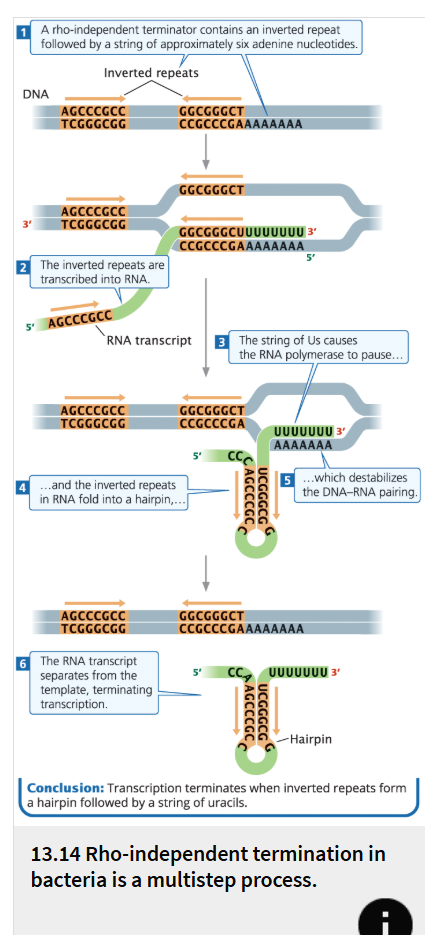
Rho-independent terminators
(also known as intrinsic terminators) are able to cause the end of transcription in the absence of the rho factor.
make up about 50% of all terminators in prokaryotes
have two common features.
First, they contain inverted repeats, which are sequences of nucleotides on the same strand that are inverted and complementary.
When these inverted repeats are transcribed into RNA and bind to each other, a hairpin forms
Second, in rho-independent terminators, a string of seven to nine adenine nucleotides follows the inverted repeat in the template DNA.
The transcription of these adenines produces a string of uracil nucleotides after the hairpin in the transcribed RNA.
rho utilization (rut) site
serves as a binding site for the rho factor
polycistronic mRNA
RNA molecule that is made up from single group of transcribed genes
produced when a single terminator is present at the end of a group of several genes that are transcribed together, instead of each gene having its own terminator.
Polycistronic mRNA does occur in some eukaryotes, such as Caenorhabditis elegans, but it is uncommon in eukaryotes.
Acetyltransferases
add acetyl groups to amino acids at the ends of the histone proteins, which destabilize nucleosome structure and make the DNA more accessible.
transcription factors
bind to DNA sequences and affect levels of transcription.
general transcription factors
combine with RNA polymerase to form the basal transcription apparatus
basal transcription apparatus
group of proteins that assembles near the transcription start site and is sufficient to initiate minimal levels of transcription.
consists of RNA polymerase II, a series of general transcription factors, and a complex of proteins known as the mediator
The general transcription factors include TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH, in which TFII stands for “transcription factor for RNA polymerase II” and the final letter designates the individual factor.
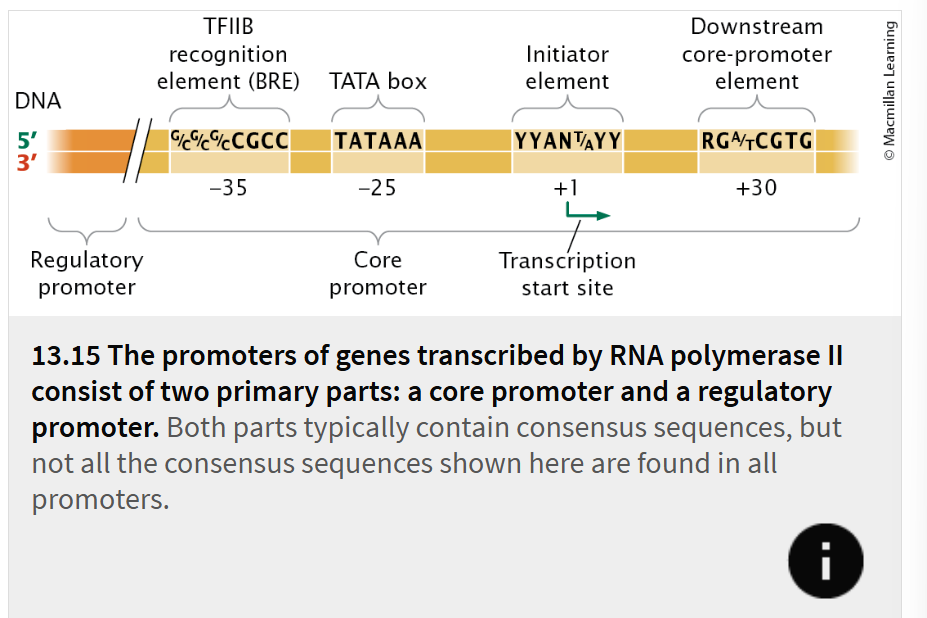
A promoter for a gene transcribed by RNA polymerase II typically consists of two primary parts:
core promoter
regulatory promoter
core promoter
located immediately upstream of the gene and is the site to which the basal transcription apparatus binds.
The core promoter typically includes one or more consensus sequences. One of the most common of these sequences is the TATA box, which has the consensus sequence TATAAA and is located −25 to −30 bp upstream of the start site.
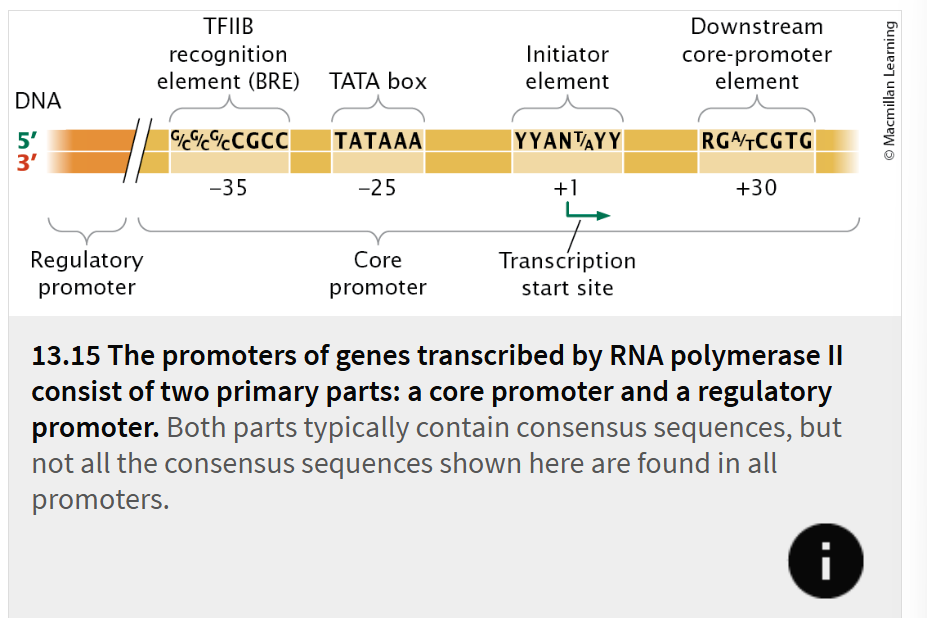
regulatory promoter
located immediately upstream of the core promoter (
A variety of different consensus sequences can be found in regulatory promoters, and they can be mixed and matched in different combinations
Transcription factors bind to these sequences and, either directly or indirectly, make contact with the basal transcription apparatus and affect the rate at which transcription is initiated.
enhancer
Sequence that stimulates maximal transcription of distant genes; affects only genes on the same DNA molecule (is cis acting)
contains short consensus sequences
is not fixed in relation to the transcription start site
can stimulate promoters in its vicinity
may be upstream or downstream of the gene.
The function of an enhancer is independent of sequence orientation.
internal promoters
Promoter located within the sequences of DNA that are transcribed into RNA.
TATA-binding protein (TBP)
recognizes and binds to the TATA box.
The TATA-binding protein binds to the minor groove of the DNA double helix and straddles the DNA like a molecular saddle, bending the DNA and partly unwinding it.
Rat1
cleaves off excess rna nucleotides during termination
5’ to 3’ exonuclease activity
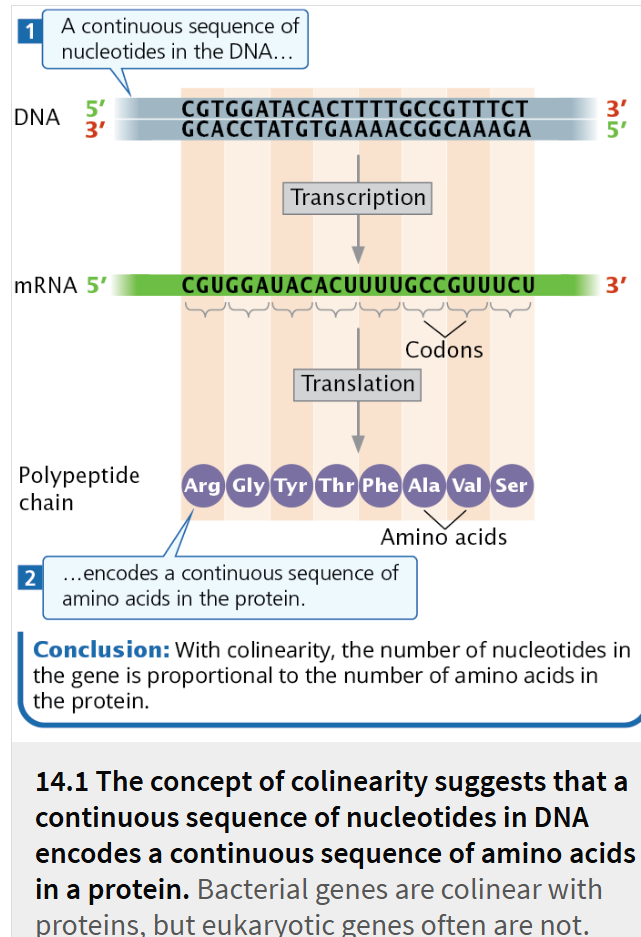
colinear
there is a direct correspondence between the nucleotide sequence of DNA and the amino acid sequence of a protein
The concept of colinearity suggests that the number of nucleotides in a gene should be proportional to the number of amino acids in the protein encoded by that gene
this concept is true for genes found in bacterial cells and in many viruses,
although these genes are slightly longer than would be expected if colinearity were strictly applied because the mRNAs encoded by the genes contain sequences at their ends that do not specify amino acids.
exons
RNA coding regions
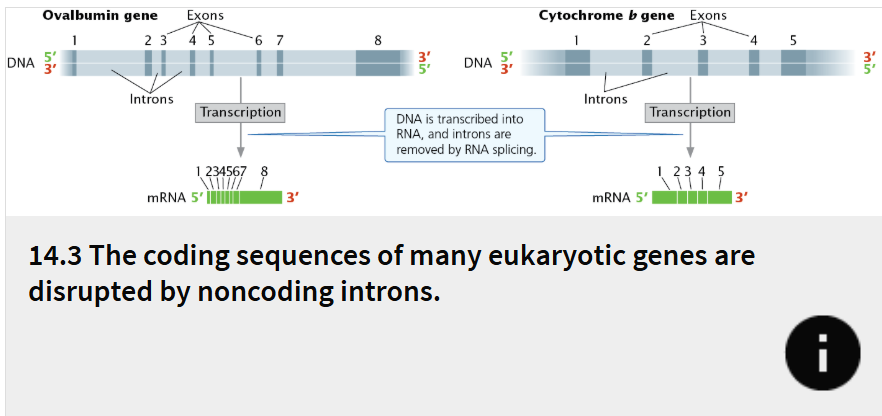
introns
noncoding regions called intervening sequences

Group I introns
found in some genes of bacteria, bacteriophages, and eukaryotes, are self-splicing: they can catalyze their own removal
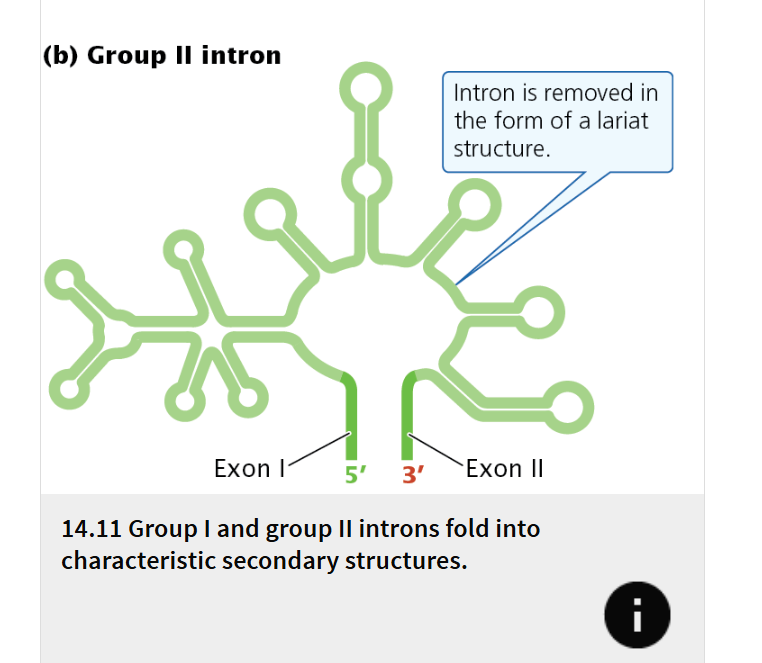
Group II introns
present in some genes of mitochondria, chloroplasts, archaea, and a few bacteria
they are also self-splicing, but their mechanism of splicing differs from that of the group I introns.
nuclear pre-mRNA introns
are the best studied
they include introns located in the protein-encoding genes of the eukaryotic nucleus.
The splicing mechanism by which these introns are removed is similar to that of the group II introns, but nuclear introns are not self-splicing
their removal requires a large structure called a spliceosome that includes small nuclear RNAs (snRNAs, discussed in Section 14.2) and a number of proteins.
transfer RNA introns
found in tRNA genes of bacteria, archaea, and eukaryotes, are removed by yet another splicing mechanism, one that relies on enzymes to cut and reseal the RNA.
codon
set of three nucleotides that specifies the amino acid in an mRNA molecule
5’ untranslated region (5’ UTR)
sometimes called the leader, is a sequence of nucleotides at the 5′ end of the mRNA that does not encode any of the amino acids of a protein.
In bacterial mRNA, this region contains a consensus sequence (AGGAGG) called the Shine–Dalgarno sequence
Shine–Dalgarno sequence
AGGAGG
serves as the ribosome-binding site during translation
it is found approximately seven nucleotides upstream of the first codon translated into an amino acid (called the start codon).
complementary to a sequence found in one of the RNA molecules that make up the ribosome, and it pairs with that sequence during translation.
Eukaryotic mRNA has no equivalent RNA-binding consensus sequence in its 5′ untranslated region.
In eukaryotic cells, ribosomes bind to a modified 5′ end of mRNA, as discussed later in this section.

protein-coding region
comprises the codons that specify the amino acid sequence of the protein.
The protein-coding region begins with a start codon and ends with a stop codon.
3′ untranslated region (3′ UTR)
sometimes called the trailer, a sequence of nucleotides at the 3′ end of the mRNA that is not translated into amino acids.
The 3′ UTR affects the stability of mRNA and helps regulate the translation of the mRNA protein-coding sequence.
5’ cap
One type of modification of eukaryotic pre-mRNA
functions in initiation of translation

poly(A) tail
A second type of modification to eukaryotic mRNA is the addition of 50 to 250 or more adenine nucleotides at the 3′ end
not encoded in the DNA but are added after transcription in a process termed polyadenylation
Processing of the 3′ end of pre-mRNA requires sequences (collectively termed the polyadenylation signal) located both upstream and downstream of the site where cleavage occurs.
The consensus sequence AAUAAA (called the poly(A) consensus sequence) is usually 11 to 30 nucleotides upstream of the cleavage site and determines the point at which cleavage will take place.
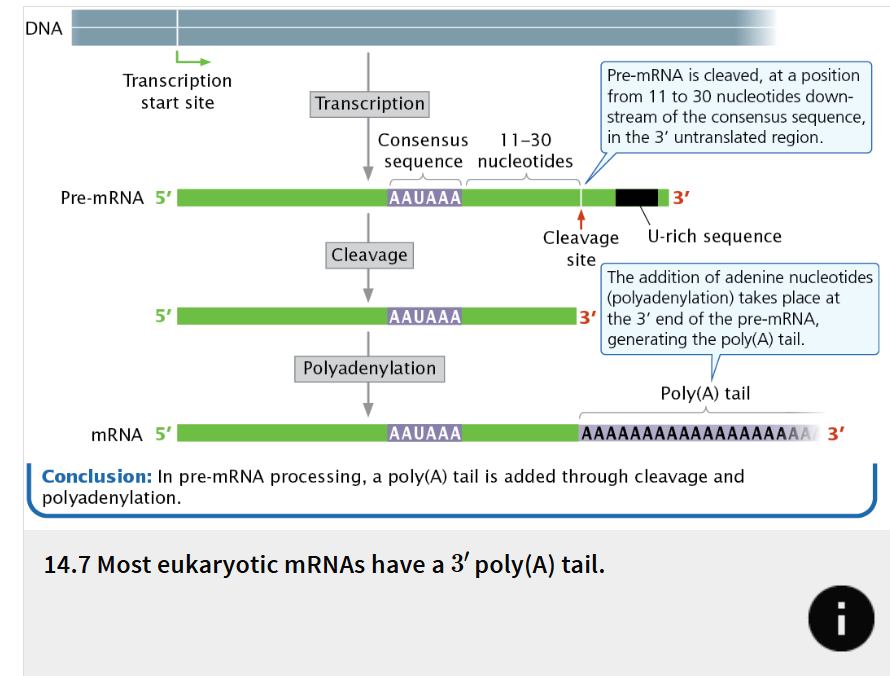
Purpose of poly(A) tail and 5’ cap
confer stability on mRNA by protecting it from degradation by 5′ and 3′ exonucleases.
The stability conferred by the poly(A) tail depends on proteins that attach to the tail, and on its length.
The poly(A) tail also facilitates attachment of the ribosome to the mRNA and plays a role in export of the mRNA to the cytoplasm.
RNA splicing
other major type of modification of eukaryotic pre-mRNA that removes introns
This modification takes place in the nucleus, before the RNA moves to the cytoplasm.
Splicing occurs co-transcriptionally, which means that transcription and splicing take place at the same time.
While transcription is continuing at the 3′ end of the growing pre-mRNA, introns may be removed from that part of the pre-mRNA that has already been transcribed
5’ splice site and 3’ splice site
these splice sites possess short consensus sequences.
Most introns in pre-mRNAs begin with GU and end with AG, indicating that these sequences play a crucial role in splicing.
Indeed, changing a single nucleotide at either of these sites prevents splicing.

Splicing requires the presence of three sequences in the intron:
(termed the “splicing code”
5’ splice site
3’ splice site
branch point
branch point
an adenine nucleotide that lies 18 to 40 nucleotides upstream of the 3′ splice site
.The sequence surrounding the branch point does not have a strong consensus.
Deletion or mutation of the adenine nucleotide at the branch point prevents splicing
spliceosome
large structure where spli
consists of five RNA molecules and almost 300 proteins.
The RNA molecules are small nuclear RNAs (snRNAs) ranging in length from 107 to 210 nucleotides
each snRNA associates with proteins to form a small nuclear ribonucleoprotein particle (snRNP).
The spliceosome is composed of five snRNPs (U1, U2, U4, U5, and U6) and other proteins not associated with an snRNA.cing takes place
lariat
Loop-like structure created in the splicing of nuclear pre-mRNA when the 5′ end of an intron is attached to the branch point.

trans-splicing
Process of splicing together exons from two or more pre-mRNAs.
self-splicing introns
Some introns are self-splicing—they possess the ability to remove themselves from an RNA molecule without the aid of enzymes or other proteins.
These self-splicing introns fall into two major categories: group I introns and group II introns
group I introns
Group I introns are found in a variety of genes, including some rRNA genes in protists, some mitochondrial genes in fungi, and even some bacterial and bacteriophage genes.
Although the lengths of group I introns vary, all of them fold into a common secondary structure with nine hairpins (stem-loop structures), which are necessary for splicing.
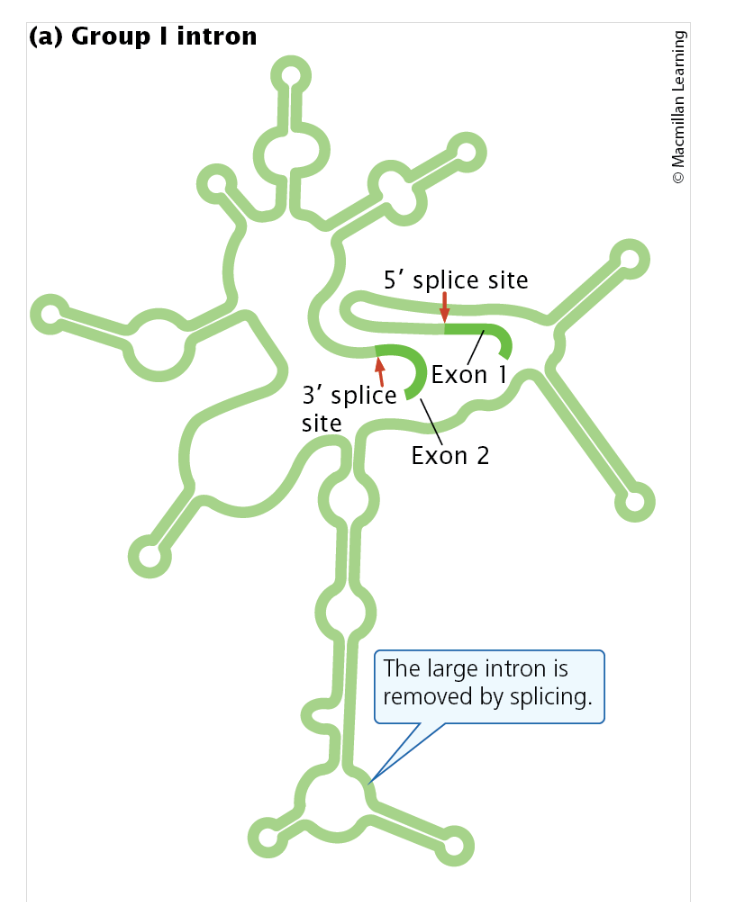
Group II introns
present in genes of bacteria, archaea, and eukaryotic organelles, also have the ability to self-splice.
All group II introns also fold into secondary structures
The splicing of group II introns is accomplished by a mechanism that has some similarities to the spliceosome-mediated splicing of nuclear pre-mRNA introns, and splicing generates a lariat structure.
Because of these similarities, group II introns and nuclear pre-mRNA introns are thought to be evolutionarily related: nuclear introns probably evolved from self-splicing group II introns and later adopted the proteins and snRNAs of the spliceosome to carry out the splicing reaction.
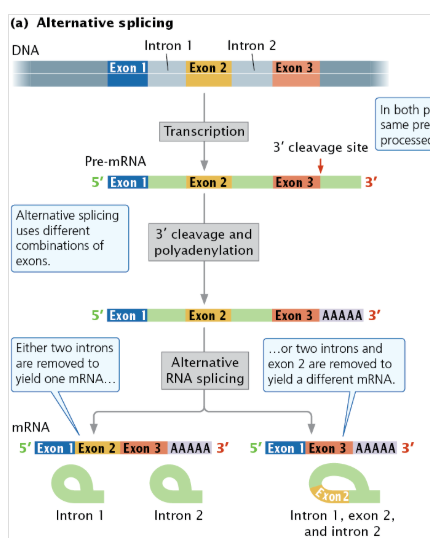
alternative processing pathway
One of several pathways by which a single pre-mRNA can be processed in different ways to produce alternative types of mRNA.
results in the production of different proteins from the same DNA sequence
alternative splicing
type of alternative processing
the same pre-mRNA can be spliced in more than one way to yield different mRNAs that are translated into different amino acid sequences and thus different proteins
multiple 3’ cleavage sites
type of alternative processing
The presence of more than one 3′ cleavage site on a single pre-mRNA, which allows cleavage and polyadenylation to take place at different sites, producing mRNAs of different lengths.

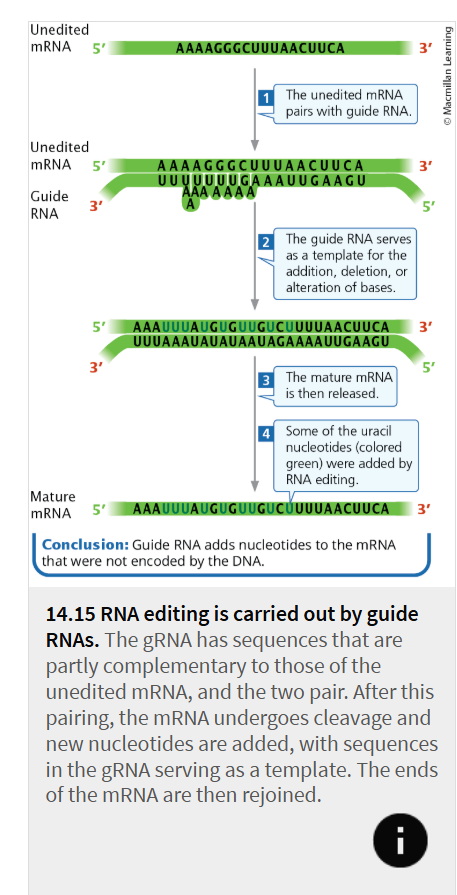
RNA editing
the coding sequence of an mRNA molecule is altered after transcription, so that the translated protein has an amino acid sequence that differs from that encoded by the gene.
guide RNAs (gRNAs)
modifies sequences to produce an edited mRNA molecule
contains sequences that are partly complementary to segments of the unedited mRNA, and the two molecules undergo base pairing at these sequences
After the mRNA is anchored to the gRNA, the mRNA undergoes cleavage and nucleotides are added, deleted, or altered according to the template provided by the gRNA.
modified bases
Any of several rare bases found in some RNA molecules. Such bases are modified forms of the standard bases (adenine, cytosine, guanine, and uracil).
but tRNAs have nucleotides containing additional bases, including ribothymidine, pseudouridine (which is also occasionally present in snRNAs and rRNA), and dozens of others

tRNA-modifying enzymes
carry out chemical changes to the four standard bases after transcription to create modified bases
cloverleaf
Secondary structure with four major arms common to all tRNAs
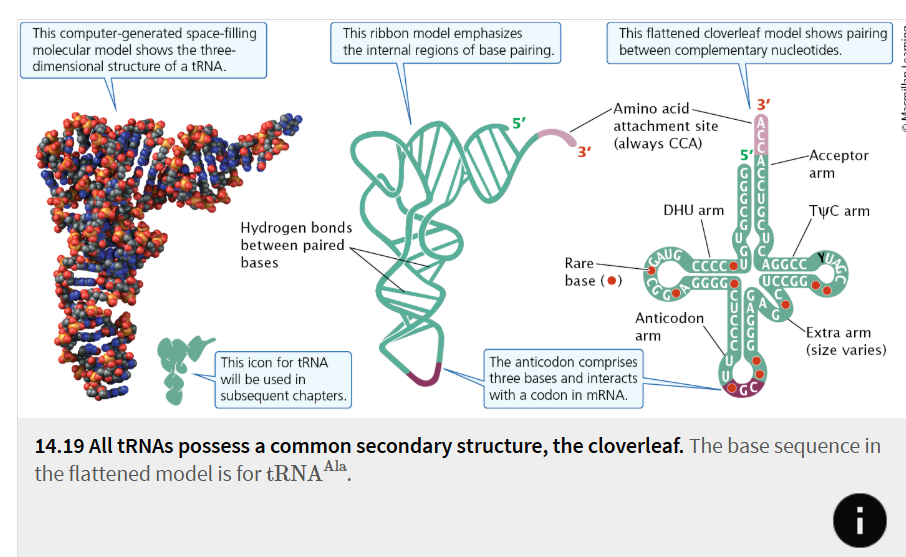
anticodon
Sequence of three nucleotides in tRNA that pairs with the corresponding codon on mRNA to ensure that the amino acids link in the correct order during translation.
A functional ribosome consists of two subunits:
a large ribosomal subunit and a small ribosomal subunit,
each of which consists of one or more RNA molecules and a number of proteins.

Svedberg (S) units
sizes of the ribosomes and their RNA components
(a measure of how rapidly an object sediments in a centrifugal field).
(It is important to note that S units are not additive: combining a 10S structure and a 20S structure does not necessarily produce a 30S structure because the sedimentation rate is affected by the three-dimensional structure of an object as well as by its mass.)
RNA interference (RNAi)
a powerful and precise mechanism used by eukaryotic cells to limit the invasion of DNA sequences (from viruses and transposons) and to censor the expression of their own genes.
triggered by double-stranded RNA molecules, which may arise in several ways:
by the transcription of inverted repeats into an RNA molecule that then base pairs with itself to form double-stranded RNA
by the simultaneous transcription of two different RNA molecules that are complementary to each other and that pair, forming double-stranded RNA
or by infection by viruses that make double-stranded RNA

RNA-induced silencing complex (RISC)
Complex of a small interfering RNA (siRNA) or microRNA (miRNA) with proteins that can cleave mRNA, leading to degradation of the mRNA or repression of its translation.
long noncoding RNAs (IncRNAs)
A class of relatively long RNA molecules found in eukaryotes that do not code for proteins; they provide a variety of functions, including regulation of gene expression.
enhancer RNAs (eRNAs)
noncoding
are transcribed from enhancer sequences and may play a role in regulating the expression of protein-encoding genes, perhaps by facilitating the looping of DNA between enhancers and promoters