BIOL 3301: Chapter 17 Non-Coding RNAs
1/46
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
47 Terms
noncoding RNAs ncRNAs
some genes don’t code polypeptides, but are transcribed into ?
est 1000s-10,000s ?’s in humans
perform wide array of cellular fx’s in bacteria, archaea, eukaryotes
in most cell types, ? are more abundant than mRNAs
in a typical human cell, only about 20% of transcription involves making mRNAs, whereas 80% focuses on making ?
ncRNAs
in most cell types, ? are more abundant than mRNAs
in a typical human cell, only about 20% of transcription involves making mRNAs, whereas 80% focuses on making ?
CRISPR Cas system
system in some prokaryotes that defends against foreign invaders
defense against bacteriophages, plasmids, transposons
ncRNAs play a key role
3 types exist
we focus on type II
CRISPR Cas system
provides bacteria with defense against bacteriophages
CRISPR locus recognized in 1993
site in prokaryotic chromosomes
contains a series of repeated sequences
Francis Mojica, Giles Vergnaud, Alexander Bolotin independently proposed it provides protection against bacteriophage infection in 2005
based on sequence analysis revealing that the CRISPR locus contains segments derived from bacteriophage DNA
Philippe Horvath and colleagues experimentally showed that the ? system provides defense against bacteriophage infection in 2007
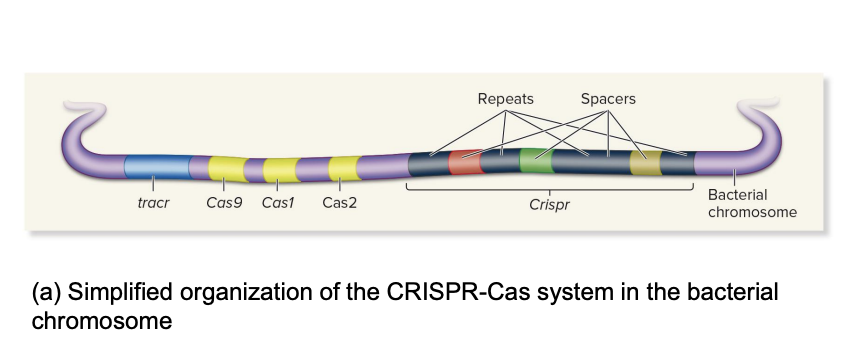
CRISPR-Cas system organization
ex., 5 genes
tracr
process pre-crRNA into usable crRNA
binds crRNA and guides Cas9 to target DNA
Cas9
“scissors”
cuts foreign DNA when guided by crRNA/tracrRNA
Cas1
work with Cas2 to recognize and insert new viral DNA pieces into CRISPR as new spacers (adaptation phase)
create genetic memory of new invaders
Cas2
“ “ but works with Cas 1
Crispr
group of Clustered, Regularly, Interspaced, Short, Palindromic Repeats
repeats
identical sequences
spaces
repeats are interspersed with spacers (short, unique sequences)
unique bits of viral DNA (memory of past infections)
CRISPR analogy
CRISPR is a library of viral mugshorts
Cas1 and Cas2 add new mugshots into CRISPR
insert new viral DNA pieces as spacers
tracrRNA + crRNA guide Cas9 to arrest the invader (cut viral DNA)
adaptive defense system
CRISPR Cas system is an ?
bacterial cell must first be exposed to an agent (e.g., bacteriophage) to elicit a response
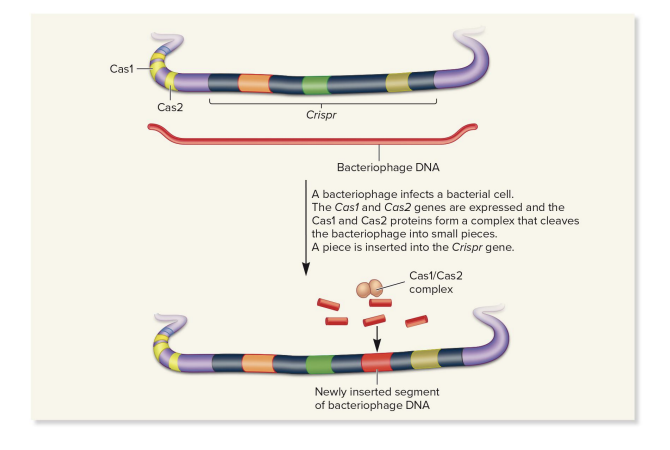
CRISPR adaptation phase
(infection first happens, invading DNA is used to adapt the cell to future infections)
Cas1 and Cas2 protein complex cleaves bacteriophage DNA
a piece of this bacteriophage DNA (typically 20-50 bp) is inserted into the Crispr locus as a new spacer
spacers in modern bacteria are from past infections
genetic “memories” of past infections
the spacer is passed to daughter cells, giving them immunity to same virus in future
CRISPR expression phase
upon re-exposure, the Crispr, tracr, and Cas9 genes are transcribed
2 noncoding RNAs (ncRNAs) are made
pre-crRNA
tracr-RNA
region of tracrRNA base pairs with repeats in pre-crRNA
the pre-crRNA is then cleaved into many small crRNAs (each contains 1 spacer)
tracrRNA-crRNA complex binds Cas9 protein thru recognition site in tracrRNA
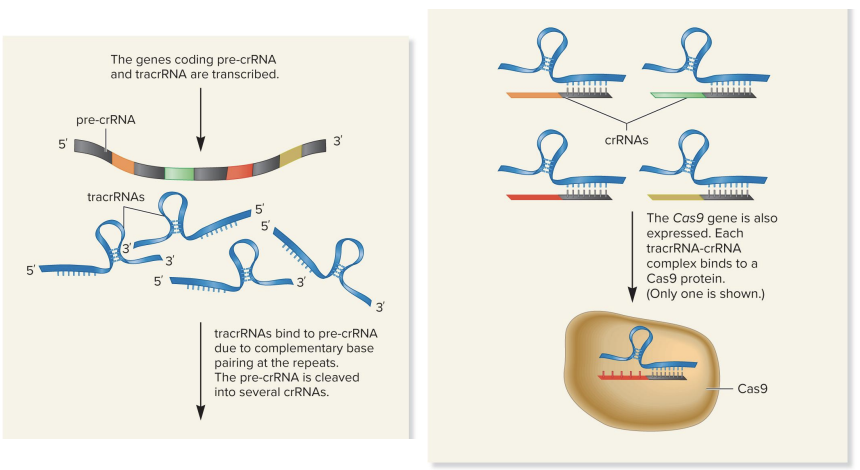
CRISPR interference phase
during reinfection
each spacer in a crRNA is complementary to one strand of the bacteriophage DNA
crRNA acts as a guide that causes the tracrRNA-crRNA-Cas9 complex to bind to that strand
Cas9 protein functions as an endonuclease→makes DOUBLE-STRANDED BREAKS in the bacteriophage DNA
bacteriophage proliferation is inhibited
gene editing via CRISPR Cas
experimentally altering a gene sequence
analysis of mutations can provide important info about normal genetic processes
mutations can rise spontaneously, or be induced by mutagens
researchers have developed techniques to make mutations within cloned DNA and also within the DNA of living cells
crispr cas technology
the system can also be used to edit genes in living cells
e.g., if you want to test the fx of a gene by “knocking it out” i.e., mutating the gene so that no functional protein is made
tracrRNA and crRNA are fused into one molecule
called the single guide RNA (sgRNA); aka guide RNA gRNA
the spacer region of the sgRNA is complementary to one of the strands of the gene to be edited (called the target gene)
sgRNA binds to Cas9 and guides it to the target gene
Cas9 then makes a double strand break in the gene
after Cas9 cuts the DNA, 2 different repair events are possible
nonhomologous end joining NHEJ
region may incur a small deletion or insertion that inactivates gene (knockout)
homologous recombination repair
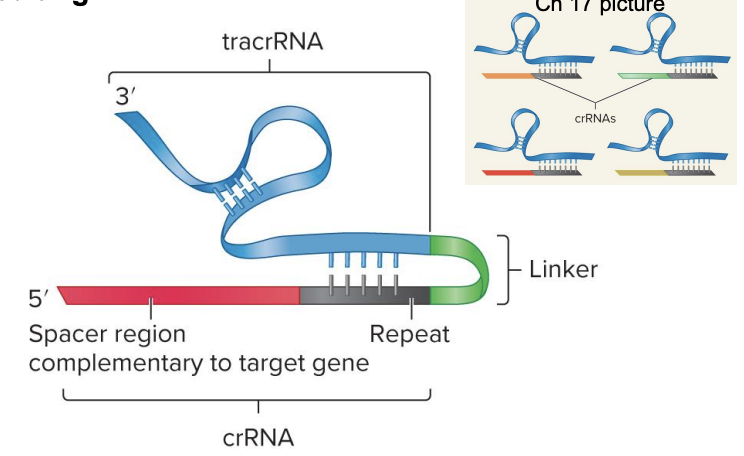
single guide sgRNA structure
crRNA + tracrRNA joined by a linker
tracrRNA helps process pre-crRNA into crRNA
recruit/guide Cas9 to target
crRNA guides Cas9 to specific viral DNA to cut
contains “spacer” sequence matching viral DNA
spacer region of sgRNA is complementary to target gene
guides Cas9 to specific location in bacteriophage DNA
nonhomologous end joining
possible repair event after cas9 cuts the DNA
joins broken ends w/o template
often causes small insertion/deletions (indels)
can inactivate gene
CRSPR Cas gene editing
Cas9+sgRNA bind target DNA sequence
sgRNA= crRNA +tracrRNA joined by a linker
sgRNA spacer is complementary to target gene
Cas9 cleaves both DNA strands, creating a double stranded break
2 options for repair
nonhomologous end joining
fast, error prone
cause small insertions/deletions
may inactivate the gene
homologous recombination repair HRR
can introduce specific point mutation
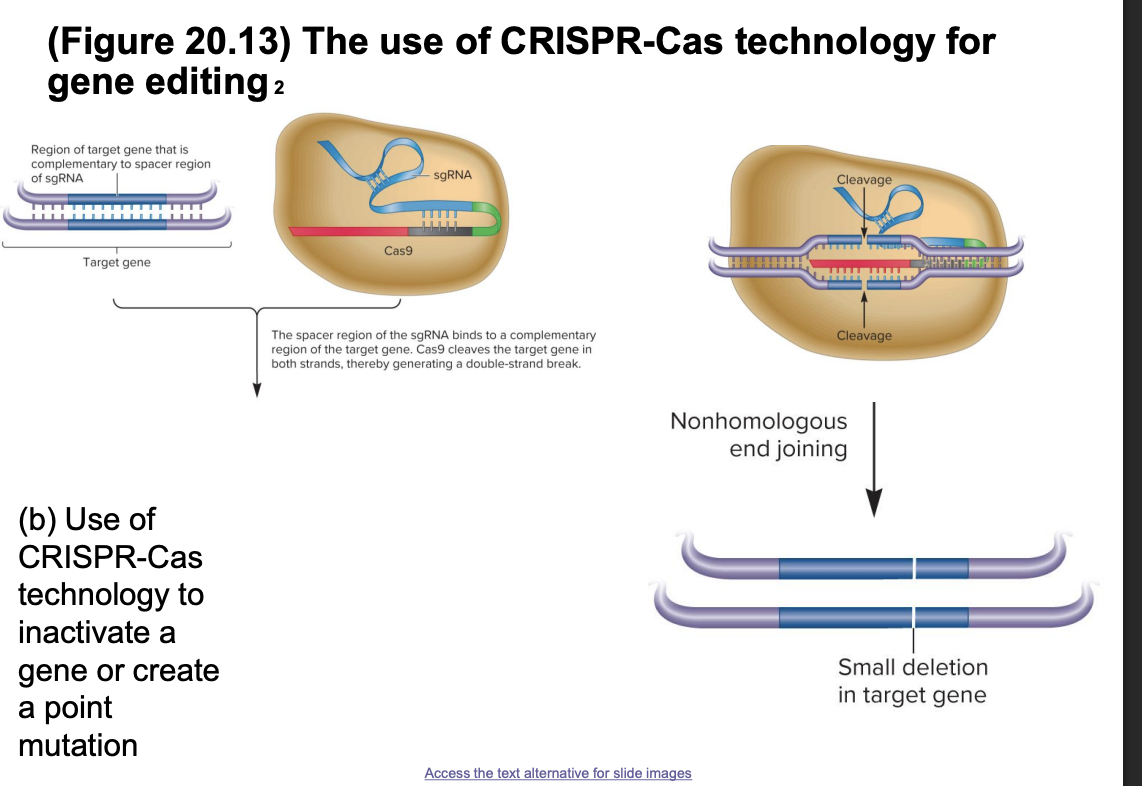
CRISPR Cas tech
can be used on different cell types and whole organisms
ex., inject a segment of DNA that encodes an sgRNA and Cas9 protein into a fertilized mouse oocyte
the 2 genes (sgRNA and Cas9 gene) are expressed in the oocyte
sgRNA-Cas9 complex would either cause a gene inactivation (NHEJ) or produce a point mutation (HRR)
ex., mouse embryos, zebrafish, frogs, adult mice, human cell lines, roundworms, variety of different plant cell species
CRISPR based therapeutics
2021: FDA approves CRISPR based treatment for sickle cell disease for clinical trials
2018: FDA approves clinical trial of CRISP based treatment for Leber congenital amaurosis (LCA), a vision loss disease
one, multiple
thousands of genetic diseases afflict ppl
many of these are the direct result of a mutation in ? gene
many involve ? genes
complex pattern of inheritance
ex., diabetes, asthma, mental illness
genetic diseases
study of this provides insight regarding our traits
ex., analyzing ppl with hemophilia
researchers have ID’ed genes that participate in blood clotting
thousands of human diseases have a genetic basis
we focus on those resulting from mutations in single genes
the occurrence of these diseases in family pedigrees often obey simple Mendelian inheritance patterns
family pedigrees
genetic diseases here often obey simple Mendelian inheritance patterns
genes
several observations are consistent w/ idea that a disease is caused, at least in part, by ?
when an individual exhibits a disease, the disorder is more likely to occur in genetic relatives than in the general poulation
identical twins share the disease more often than fraternal twins
concordance is 1 for identical twins
the disease does not spread to individuals sharing similar environmental situations
different populations tend to have different frequencies of the disease
the disease tends to develop at a characteristic age
many genetic disorders exhibit a specific age of onset
the human disorder resembles a genetic disorder that has a genetic basis in another mammal
a correlation is observed btw a disease and a mutant human gene or chromosomal alteration
monozygotic twins MZ
formed from same sperm and egg
identical twins
concordance of disorder is 1
%age of twin pairs in which both twins exhibit the disorder or trait
dizygotic twins
formed from separate pairs of sperm and egg
fraternal twins
concordance
the degree to which a disease is inherited
refers to the percentage of twin pairs in which both twins exhibit the disorder or trait
1 for identical twins
age of onset
many genetic disorders exhibit a specific ?
disease tends to develop at a characteristic age
pedigree analysis
inheritance patterns of human diseases may be determined via ?
the pattern of inheritance of a human disorder that is caused by a mutation in a single gene can be deduced by analyzing human ?
to use this method, a geneticist must obtain data from large ? with many affected individuals
autosomal recessive inheritance
tay sachs disease TSD
affected individuals appear healthy at birth, but develop neurodegenerative symptoms at 4-6 months
cerebral degeneration, blindness, and loss of motor function
TSD patients typically die at 3-4 years of age
TSD is 100x more frequent in Ashkenazi (eastern Europe) Jewish populations than in others
result of a mutation in the gene that codes the enzyme hexosaminidase A (hexA) that breaks down certain lipids in neurons
an excessive accumulation of this lipid in cells of the CNS causes the neurodegenerative symptoms
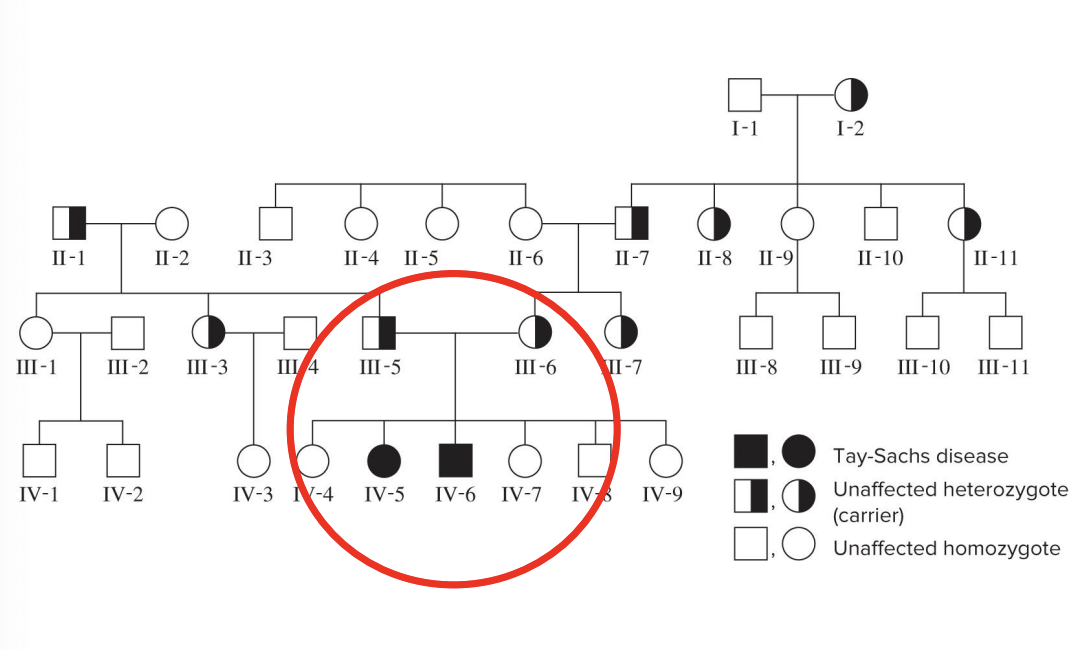
autosomal recessive inheritance
4 common features
an affected offspring will have 2 unaffected parents
when 2 unaffected heterozygotes have children, the avg percentage of affected children is 25%
2 affected individuals will have 100% affected children
the trait occurs with the same frequency in females and males
autosomal recessive
typically disorders that involve defective enzymes
the heterozygote carrier has 50% of the functional enzyme
sufficient for a healthy (unaffected) phenotype
hundreds of genetic diseases are inherited in this manner
in many cases, the mutant genes responsible have been ID’ed and characterized
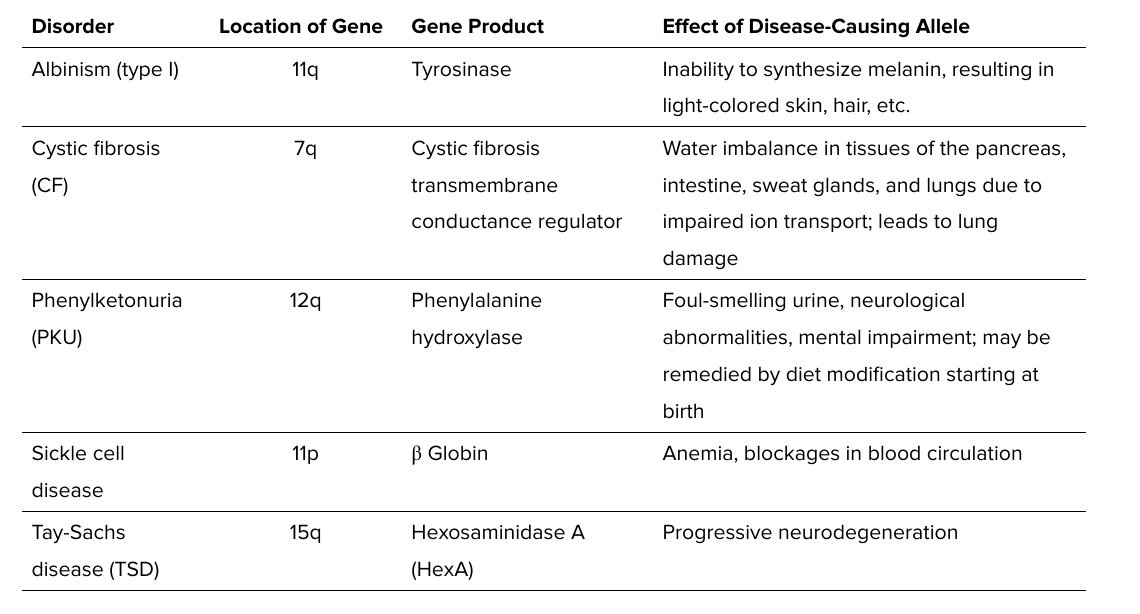
ex.,
albinism (tyrosinase)
cystic fibrosis (CFTR- a chloride transporter)
phenylketonuria PKU (phenylalanine hydroxylase)
sickle cell disease (beta globin)
tay sacs (hexosaminidase A)
autosomal recessive
equal #s M and F affected
affected kids have unaffected parents
unaffected carriers have about 25% affected kids
2 affected parents have 100% affected kids
autosomal dominant inheritance
huntington disease HD
major sx
degeneration of certain neurons in the brain
lads to personality changes, dementia, and early death (usually in middle age)
result of mutation in gene encoding protein huntingtin
mutation adds a polyglutamine tract to the protein
causes an aggregation of the protein in neurons

autosomal dominant inheritance
affected offspring usually has one or both affected parents
can be altered by reduced penetrance
affected individual with only 1 affected parent is expected to produce avg 50% affected offspring
2 affected, heterozygous individuals will have (on avg) 25% unaffected offspring
trait occurs with the same frequency in both M and F
often the homozygote is more severely affected with the disorder
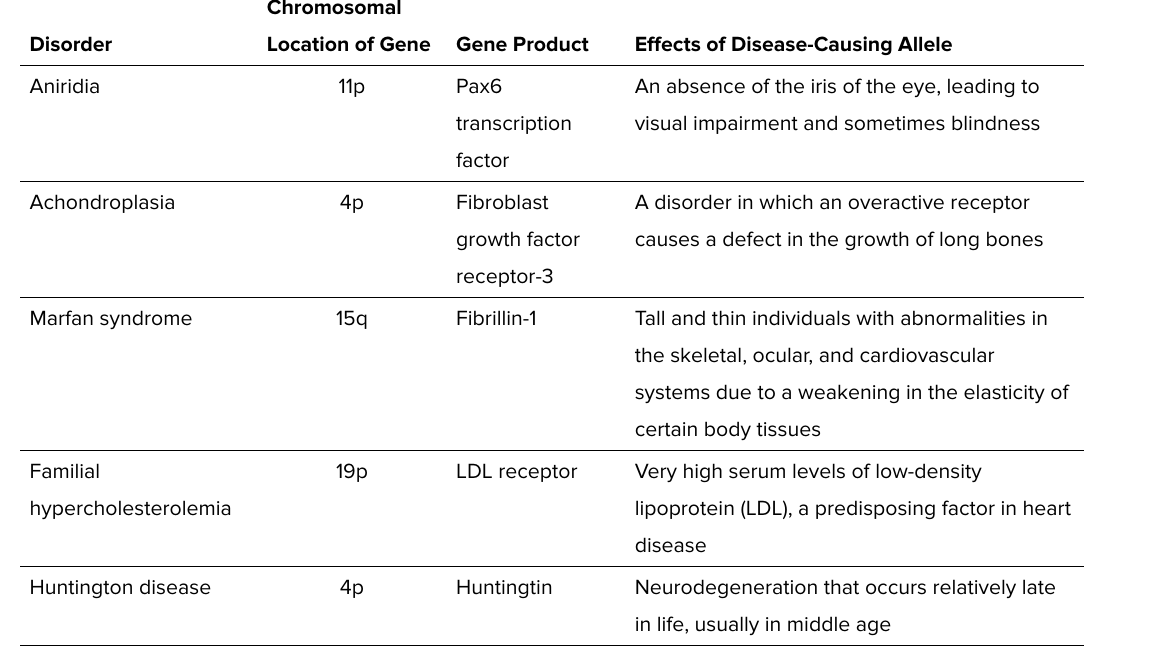
autosomal dominant disorders
3 common explanations
haploinsufficiency
heterozygote has 50% of the functional protein
not sufficient for a healthy (unaffected) phenotype
gain of function mutations
mutation changes protein so it gains a new function
dominant negative mutations
mutant gene product acts antagonistically to the wild type gene product
autosomal dominant disorders
aniridia
haploinsufficiency
achondroplasia
gain of fx
marfan syndrome
dominant negative
familial hypercholesterolemia
autosomal dominant
usually appears every generation
usually affected individuals have about 50% affected offspring
affected individuals have at least 1 affected parent
same frequency of the trait in M and F
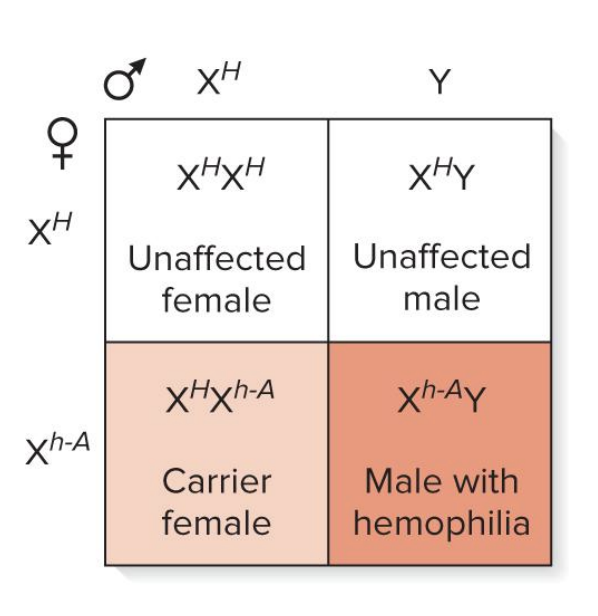
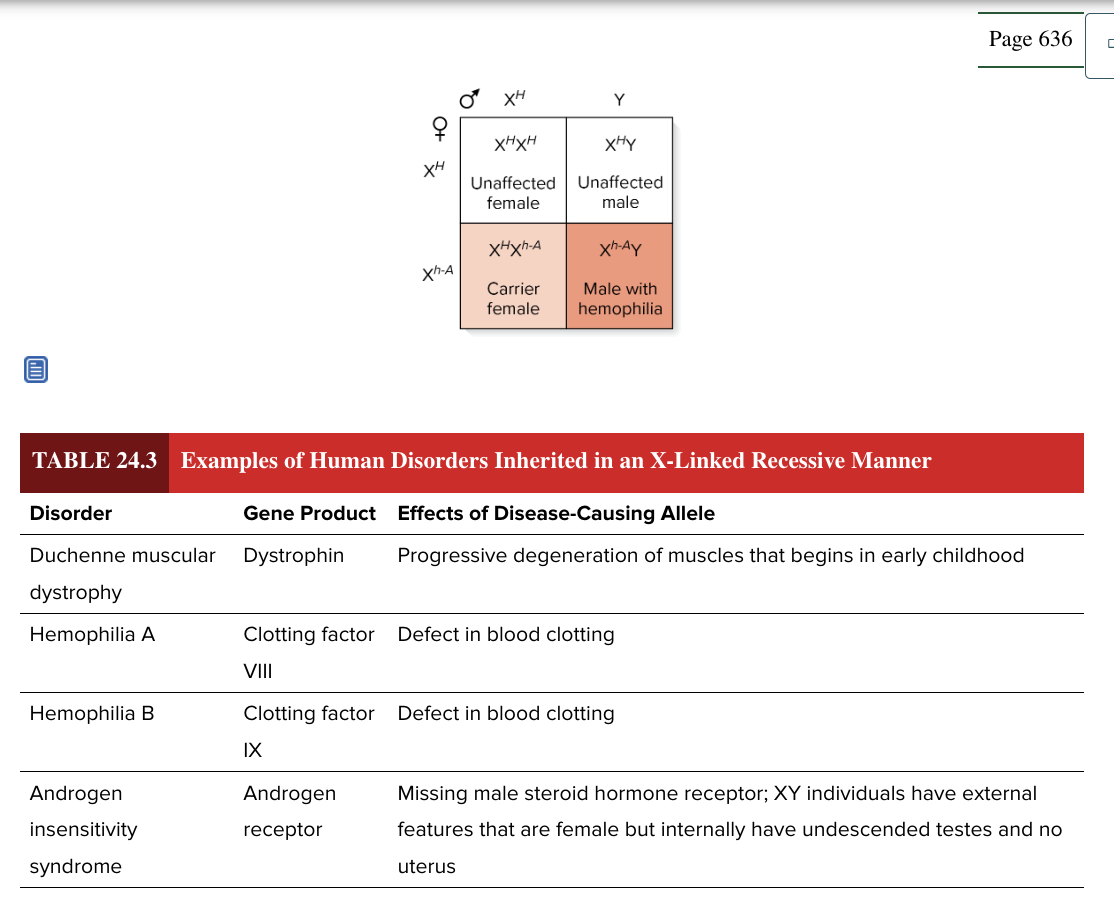
x linked recessive
this type of inheritance poses a special problem for males
males have only 1 copy of X linked genes (hemizygous)
a female heterozygous for an X linked recessive allele will pass this trait to 50% of her male offspring
x linked recessive
hemophilia
major sx
blood can’t clot properly when a wound occurs
for hemophiliacs, common accidental injuries pose a threat of severe internal or external bleeding
hemophilia A (classical hemophilia)
caused by defect in ? gene that encodes a clotting protein called factor VIII
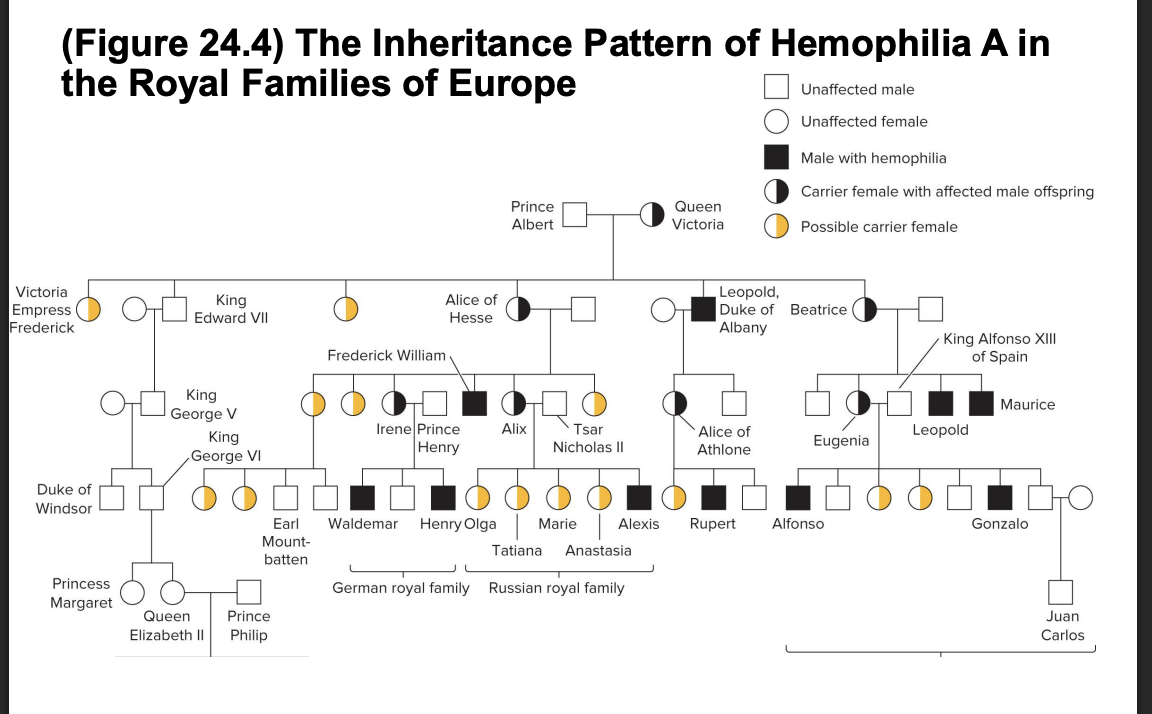
“royal disease”
affected many members of european royal families
x linked recessive
3 common features
males more likely to exhibit the trait
females with affected male offspring often have male sibling or a male parent who are affected with the same trait
the female offspring of affected males will produce (on avg) 50% affected male offspring
x linked recessive
duchenne muscular dystrophy
hemophilia A
hemophilia B
androgen insensitivity syndrome
x linked recessive
males way more affected
mother usually unaffected
females with affected fathers have about 50% affected offspring
females with affected sons usually have affected brothers
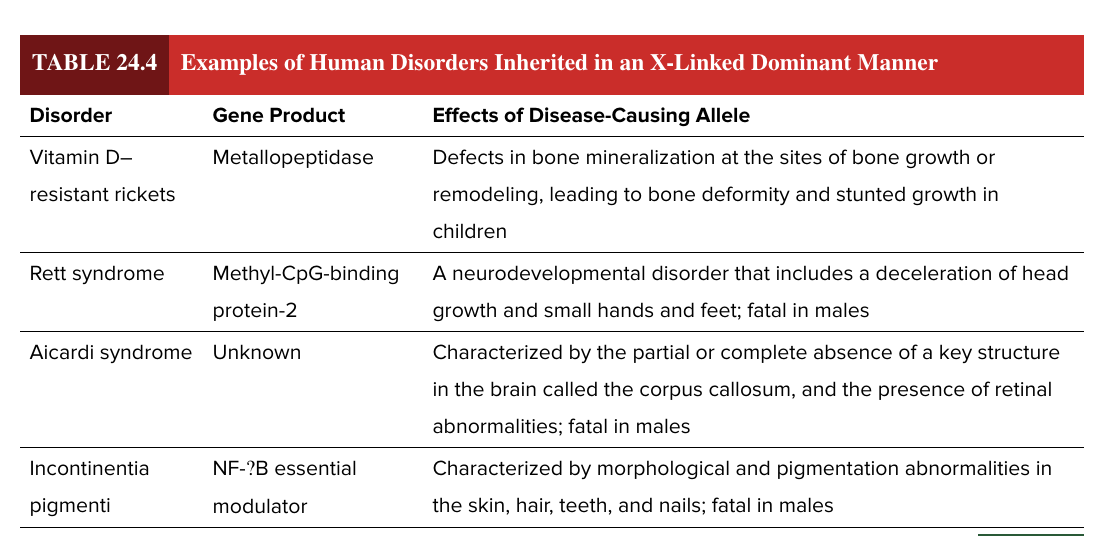
x linked dominant
rare
males often more severely affected
females may be less affected due to WT copy of the other X chromosome
F more likely to exhibit the trait when it is lethal to males
affected F have a 50% chance of passing the trait to female offspring
x linked dominant
vitamin D resistant rickets
rett syndrome
aicardi syndrome
incontinentia pigmenti
x linked dominant
rare
males usually more severely affected
if trait is lethal to males, then females show the trait more often (since there are not males that have the dominant allele)
affected females pass the trait to 50% of female offspring
pre-birth genetic testing
amniocentesis
fetal cells obtained from amniotic fluid
chorionic villus sampling
fetal cells obtained from the chorion (fetal part of placenta)
can be performed earlier during the pregnancy than amniocentesis
poses a slightly greater risk of miscarriage
noninvasive prenatal testing
collect blood sample from pregnant F and analyzing cell-free DNA
preimplantation genetic diagnosis PGD
conducted before pregnancy even occurs
genetic testing of embryos hat have been produced by IVF
IVF combines sperm + egg outside mom’s body
1 or 2 cells removed at 8 cell stage
tests conducted to check for problems
molecular tests can check for particular gene defects
chromosome composition can be checked
decision made whether or not to implant
raises ethical q’s
preimplantation genetic diagnosis PGD
conducted before pregnancy even occurs
genetic testing of embryos hat have been produced by IVF
IVF combines sperm + egg outside mom’s body
1 or 2 cells removed at 8 cell stage
tests conducted to check for problems
molecular tests can check for particular gene defects
chromosome composition can be checked
decision made whether or not to implant
raises ethical q’s
method of genetic testing prior to birth
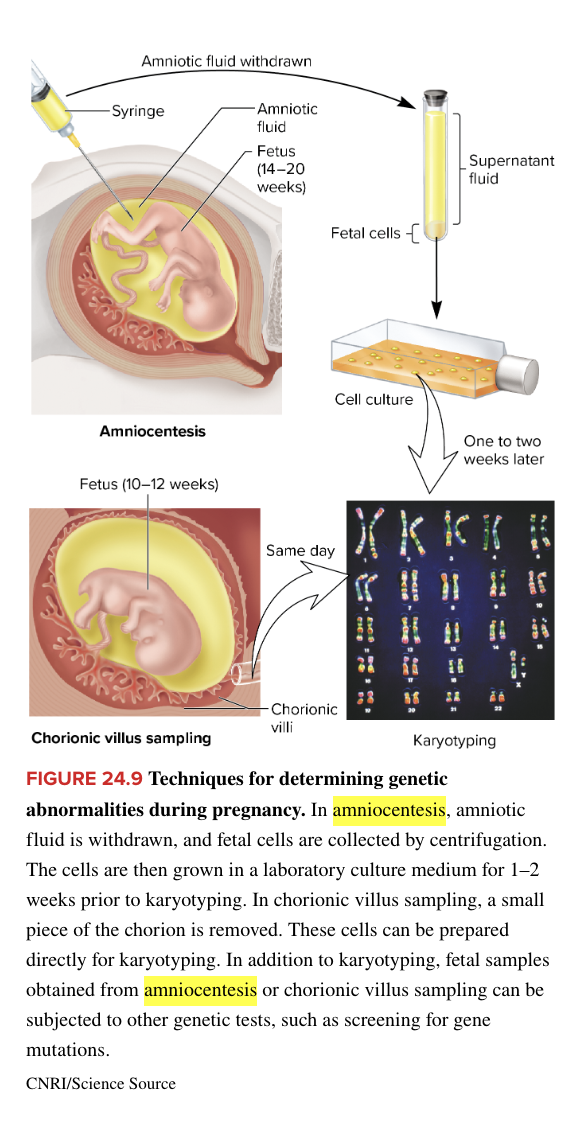
amniocentesis
fetal cells obtained from amniotic fluid
determine number of chromosomes per cell and whether changes in chrom structure have occurred
method of genetic testing prior to birth
chorionic villus sampling
fetal cells obtained from the chorion (fetal part of placenta)
can be performed earlier during the pregnancy than amniocentesis
poses a slightly greater risk of miscarriage
method of genetic testing prior to birth