L4 Protein Folding, Missfolding and Degradation
1/44
Earn XP
Description and Tags
Lecture 4 September 4th
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
45 Terms
Motif
combination of two or more secondary structures that form distinct 3D structure found in multiple proteins
is associated to a particular function
smaller than a domain
Domains
a distinct region of the protein structure
can represent a particular function, structure or refer to the spatial relationship with the rest of the protein
unstructured regions (in DNA)
regions in DNA without a fixed structure
potentially more flexible
this is in addition to the regions with the secondary structure
this is also the case in most proteins
how may domains does DNA contain ? what can we say about the motif(s) in each
4 domains
each can contain multiple motifs
Domain IV (DnaA) characteristics
Mediates DNA binding
contains two motifs that allow the interaction w specific seqs on DNA
a helix-turn-helix
a basic loop
Domain III (DnaA) characteristics
Mediates AAA ATPase
contains motifs for binding and hydrolysis of ATP
walker A
walker B
contains also other motifs and multiple other functions
what is the hierarchy of protein structure (explained through protein folding)
polypeptide folds regions w secondary structure
secondary structure elements grp into motifs
… then into domains
finally tertiary structure is acquired
how can each AA residue rotate on its axis
at aa residue portion of the polypeptide, axis of the backbone can rotate at the bonds connecting the alpha Carbon (Calpha) to the carbonyl and amide group
rotation is still limited by steric constraints imposed by the backbone and side chains
why does proline confer special conformation to proteins ?
because 5-7% of the peptide bonds with proline acquire a cis-configuration
Proline is the only naturally occurring amino acid that can exist in both cis and trans conformations
which configuration do most proteins favor and what does it result in
trans configuration (relative to the peptide backbone)
=> results in a linear backbone
trans configuration
functional groups on opposite side of plane
cis configuration
functional groups on same side of plane
isomerization
chem process by which a compound is transformed into any of its isomeric forms
Can isomerization between cis and trans proteins occur spontaneously in proline?
yes but it is slow
Peptidyl-proline isomerases (PPlases)
enzymes that speed up the process during folding of conversion of peptide bonds involving proline between their cis and trans configurations
what are results of isomerization of a single proline
can dramatically change protein structure and affect its activity
What does the combination of many small changes in the orientation of proteins despite the limitations in rotation at each aa residue?
proteins can twist and turn
how long is the process of folding?
it is very rapid — micro s to milli s
can proteins fold on their own? what would this indicate
yes some can (test tube experiments)
demonstrates that info for structure is in the protein sequence
how are hydrophobic AAs arranged in a properly folded protein
they are burried in the core and not exposed at the surface
what are hydrophobic patches at protein surface a sign of
they are a sign of misfolding
chaperones
proteins that help guide protein folding along productive pathways by permitting partially misfolded proteins to return to proper folding pathway
they facilitate the folding of many proteins
they recognize exposed hydrophobic residues
what are 3 functions of chaperones
can fold newly made proteins
refold misfolded or unfolded proteins
dissemble potentially toxic protein aggregates that form due to protein misfolding
how do chaperones work
they work through ATP-dependent cycles of binding to and release from misfolded “client” molecules at exposed hydrophobic patches
why is it useful that chaperones block the hydrophobic patches on “client” proteins?
it keeps the folding/refolding protein isolated while productive folding events occur
what are the two types of of chaperones
chaperones
chaperonins
Hsp70 (Heat-shock Proteins) chaperone mechanism
binds to short segments of an unfolded protein (like those newly syntheiszed emerging from ribosome)
binding & hydrolysis of ATP results in conformation change of Hsp70 that are needed for it to function
i.e. for protein folding assistance
what are chaperonins?
much larger complexes than chaperones that isolate unfolded proteins
GroEL chaperonin example (5 components)
composed of two stacked rings
each composed of 7 sub-units (proteins)
each ring interacts with a seren-subunit co-chaperone that acts like a lid (this is GroES which also contains 7 subunits)
in center GroEL there are chambers where all or part of protein enters
proteins less than 60kDa in mass are captured by hydrophobic residues near the entrance of the chamber
ATP hydrolysis regulates the cycle
Protein folding in the two rings is coordinated
multiple cycles can be recquired for proper folding
what happens to irretrievably misfolded proteins?
they are marked for degradation
what is the system for protein degradation?
ubiquitin/proteasome
Poly ubiquitin '“tags” damaged or misfolded proteins for degradation
Ubiquitin
a small protein
78aa residues
8.6 kDa
what tags proteins for degradation
ubiquitylation
ubiquitin becomes covalently linked to the lysine residue of target proteins
they always bind to lysine
to mark protein for degradation
what are the steps and machinery to covalently link ubiquitin to other proteins?
a carboxyl terminus is activated
ubiquitin transferred to a ubiquitin conjugating subunit: E3 ubiquitin ligase
there are 600 E3 coded in our genomes
each w specific substrate binding
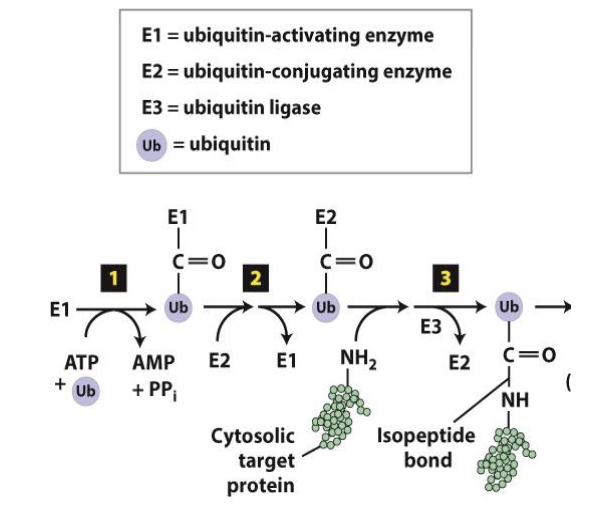
polyubiquitinynation mechanism (8 steps)
ubiquitin ligases recognize exposed hydrophobic residues
they then add multiple ubiquitins to protein (forming chain of 4 or + ubiquitins)
Polyubiquitinynated proteins are recognized by Ub receptors in the proteasome
Deubiquitinases (Dubs) hydrolyze bonds between ubiquitins to recycle them
ATPase driven auxiliary proteins unfold proteins and transports the core for degradation
once inside inner chamber, polypeptides digested into short fragmetns of 2-24aa in length
Peptide bonds of hydrophobic, acidic and basic residues are cleaved at the active site
resulting peptides further degraded into single AAs in cytoplasm
What is a proteasome
It is a large protein complex, like a cellular machine, that breaks down unwanted or damaged proteins into smaller fragments.
It is crucial for maintaining cellular health and homeostasis by eliminating proteins that are no longer needed, such as those that are misfolded or aggregated.
What does the accumulation of misfolded proteins lead to
protein aggregation
Protein aggregation
when misfolded proteins or incompletely degraded proteins iteract with each other
hiding their hydrophobic residues
forming aggregates
what leads to the formation of aggreates
high protein concentration
changes in environmental conditions
what are two ways protein aggregates can be?
amorphous
well-organized
i.e. amyloid state
IRL examples of protein aggregation
egg white contains albumin
milk contains casein
how can we see that proteon aggregation occurs in cells?
green fluorescent spot shows position of aggregates
—> we see appearance of spots in some cells as they grow suggesting the accumulation of protein aggregates
how are amyloid fibrils formed
they are formed by the generation of short segments (6-12 aa residues in length) that form long arrays or filaments of beta-sheets
each beta strand is oriented perpendicularly to the axis of the filament — two stacks twist about one another => forming photofilaments => many of the forming fibrils
where can we find amyloids and what are they markers for
they can be found in tissues
they are markers for disease
amyloid formations are associated to age but is is also more prevalent in mutant proteins
examples of neurodegenerative diseases that contain amyloids
alzheimers
parkinsons
the transmissible spongiform encephalopathy (aka “mad cow disease”)