Allosteric enzymes and Inhibitors
1/20
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
21 Terms
What is the Michaelis Menten model?
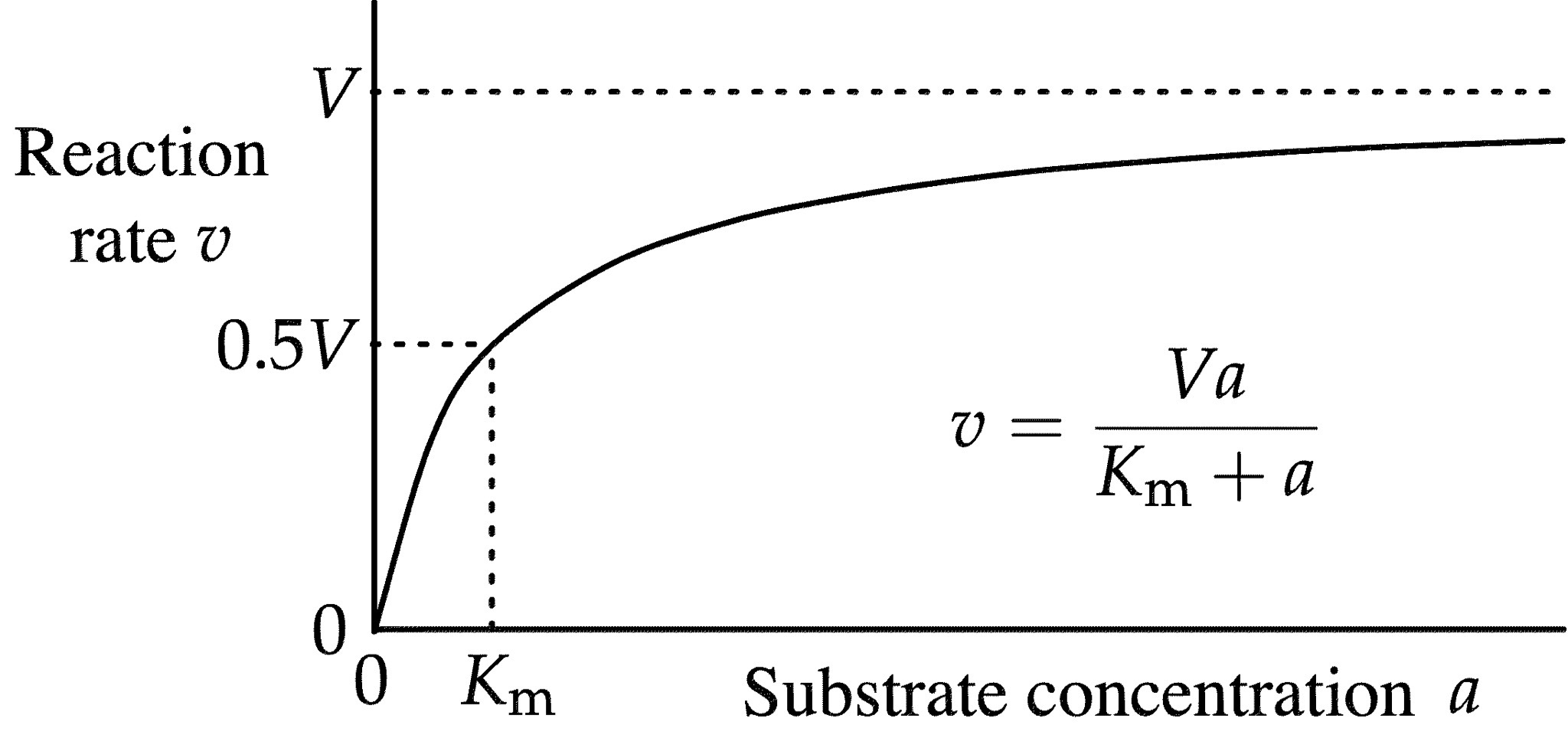
a model of enzyme kinetics that shows the relationship between the substrate concentration and the rate of an enzyme-catalysed reaction
Rate of reaction = velocity
At the initial velocity (Vo) = no product (P)
Product formation slows as we reach equilibrium. (Vmax)
Equations and
And…..
Vo = Vmax x S/Km+S
Km = K-1 + K2/K1
at low S, a doubling of S will double Vo
At a high S, increases in S will have little effect on Vo (enzyme is saturated)
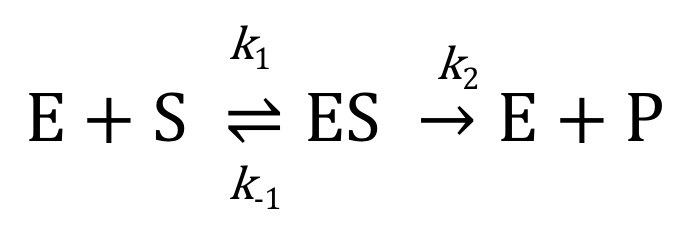
What do the K’s mean?
Km = substrate concentration at ½ Vmax (high Km = low affinity for S, low Km = high affinity for S)
K1 = association rate constant
K2 =dissociation rate constant
K3 = catalytic rate constant

Assumptions of MMM
binding is fast(ES)
Catalysis is slower and rate-limiting
At time 0, (Vo), P = 0
Enzymes exist as either free or substrate bound
Total enzyme(Et) = E+ES
Rate of formation and breakdown of ES is equal during the steady state
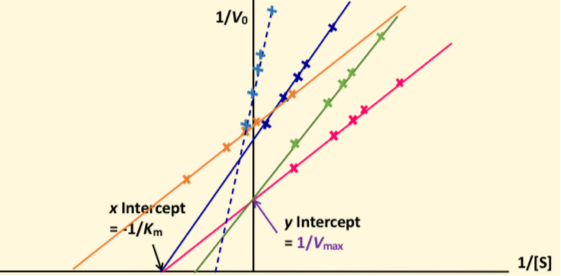
What is a lineweaver-Burke plot?
double-reciprocal graph
X= reciprocal (-1/Km) of substrate conc
Y= reciprocal (1/Vmax) of velocity
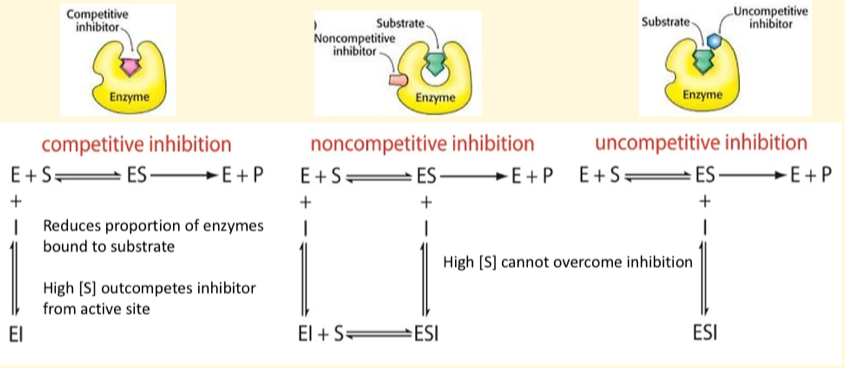
Reversible competitive inhibitors
binds reversibly to the active site
Non-covalent binding, dissociates from E
High [S] outcompetes inhibitor from active site
Effect on Vmax = no change
Effect on Km = increases
Reversible non-competitive inhibitors
Type of mixed inhibitor.
binds reversibly to site other than active site
Changes enzymes structural conformation and reduces catalytic activity
Affinity of enzyme for substrate unchanged
Non-covalent binding
Effect on Vmax = decreases
Effect on Km = no chnage
Mixed inhibitors
binds reversibly to allosteric site
Binds to E and ES complexes
If inhibitor has higher affinity for E, acts like competitive inhibitor, and decreases Km
If inhibitor has higher affinity for ES, will act uncompetitive, and Km will increase
Vmax decreases
Reversible uncompetitive inhibitors
binds to the ES complex and doesn’t allow product formation
Slope (Km/Vmax) not affected
Non-covalent binding
Effect on Vmax = decreases
Effect on Km = decreases
Irreversible inhibitors
permanently deactivate denatured enzyme
Covalent bond to active site
What do the reversible inhibitors bind to?
Competitive = only binds to free enzymes
Non-competitive = bind to free enzymes and ES complexes
Un-competitive = only binds to ES complexes
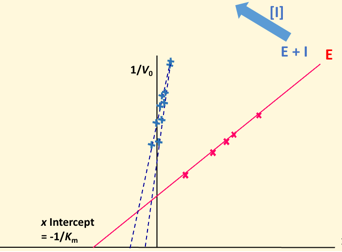
Lineweaver Burke plot - Competitive
No Vmax change, increased Km (affinity for S)
increase in Km means lower affinity for S
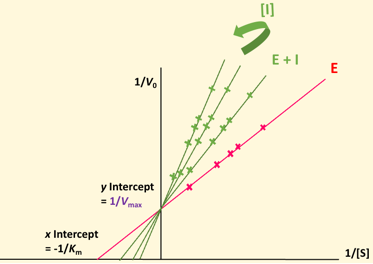
Lineweaver Burke plot - non-competitive (Mixed)
No change in Km, pivots around Km, decrease in Vmax
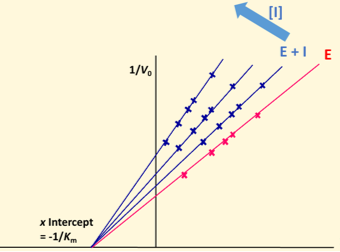
Lineweaver Burke plot - uncompetitive
Decreased Vmax and Km
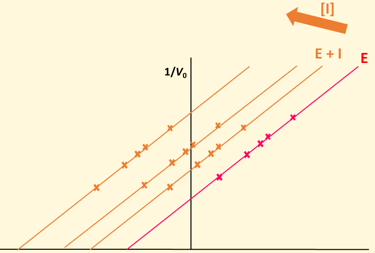
Lineweaver Burke plot - mixed
Decresed Vmax, either increase or decrease Km
What regulates enzyme activity?
allosteric control
Proteolytic activation of zymogens (inactive to active)
Transcriptional regulation → amount of E expressed and rare of mRNA degradation
Reversible covalent modification → kinases/phosphatases
Tissue specific expression of isoenzymes (different but similar AA sequences but catalyse same reactions)
Allosteric enzymes
DONT follow Michaelis Mentem kinetics
Allostery - catalytic activity altered by binding of effector/modulator to a site other than a the active site
Homotropic allosteric effector = same as substrate (multiple subunits and active sites) e.g. O2 for Haemoglobin
Heterotropic allosteric factor = regulatory molecule that is NOT the substrate, binds reversibly to active site e.g. CO2 for Haemoglobin
Homotropic effectors affect cooperativity:
Cooperativity = +ve (increased binding on active site) or -ve (decreased binding on active site)
Haemoglobin as an example of effectors
O2→ binds to catalytic sites, changes the structure so more O2 binds more easily = Homotropic effector
CO2→ binds to non-catalytic sites, decrease Haemoglobin affinity for O2
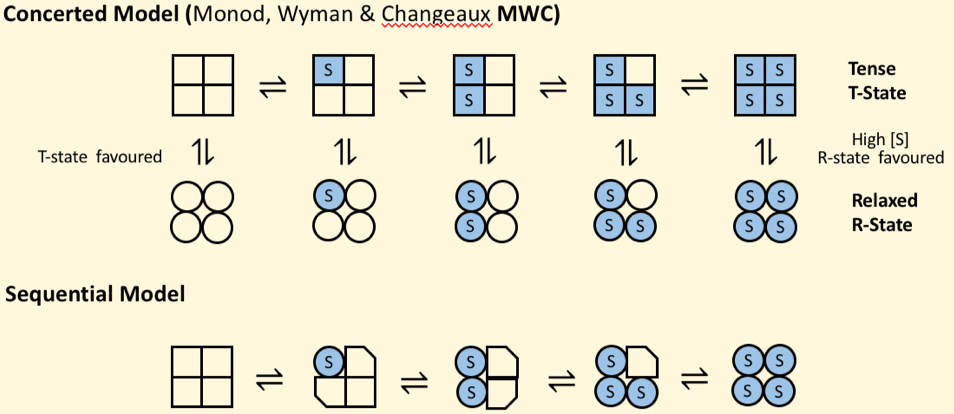
Models of Allostery
MWC model
Concerted:
subunits exist only in 2 states- T (compact) or R (relaxed)
All subunits must be in the same state
Equilibrium shifts to the R-state as more S binds
Sequential:
substrate binding increases affinity without conformational chnage of whole enzyme
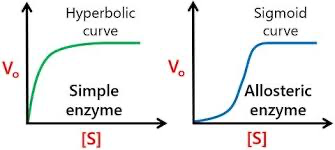
Sigmodial curve
Allosteric enzymes show cooperativity, so their activity can chnage based on substrate binding
Produces a different curve than MM kinetics
In metabolic pathways, the end product often allosterically inhibits the first enzyme (feedback intuition)
Regulation by covalent modification
Post-transcriptional modification:
phosphorylation by Ser/Thr Kinase
Reversed by action of protein phosphotases
Other:
Adenylation = transfer of adenylate from ATP
Acetylation e.g. Histones