Genetics Chapter 10: Molecular Structure of Chromosomes
1/53
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
54 Terms
Chromosome
The structures within living cells that contain the genetic material (DNA!)
Genome
The entire complement of genetic material in an organism or species.
The 4 important processes chromosomal sequences facilitate
The synthesis of RNA and cellular proteins
The replication of chromosomes
The proper segregation of chromosomes
The compaction of chromosomes so that they fit within living cells.
Protein-coding genes
Genes that carry the information to produce mRNA and that code the amino acid sequence of polypeptides.
The Nucleoid
The region of a bacterial cell where bacterial chromosomes are found.
Origin of replication
A site on a chromosome that functions as an initiation site for the assembly of several proteins that begin the process of DNA replication.
A bacterial chromosome typically is
circular
a few million nucleotides/base pairs in length
possess a few thousand different genes
has one origin of replication
has repetitive sequence dispersed in its genome
Microdomain
A loop of DNA that is found in a bacterial chromosome and is typically 10kbp in length.
Nucleoid-associated proteins
A set of DNA-binding proteins found in bacteria that facilitate chromosome compaction and organization.
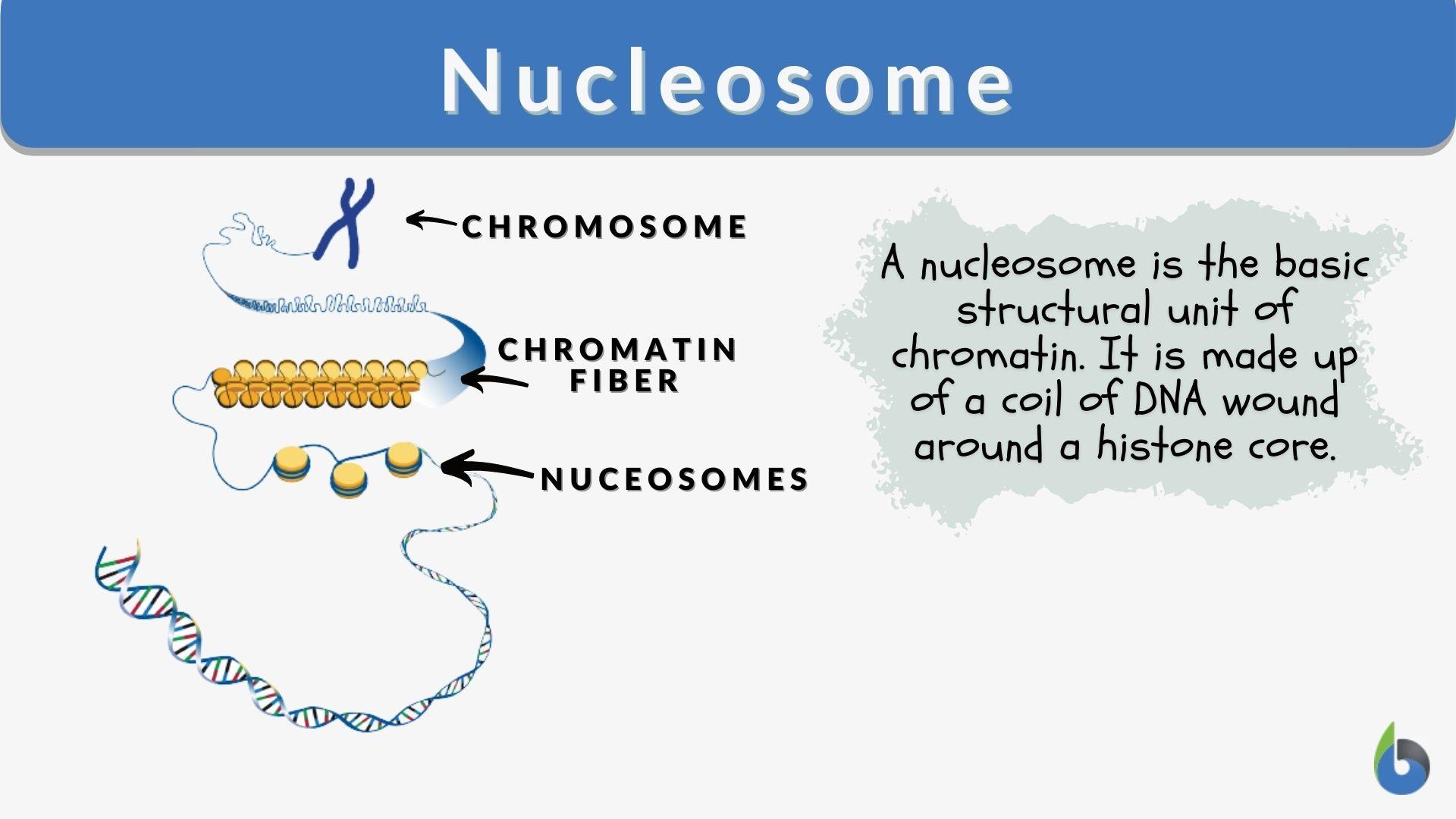
Nucleosome
The repeating structural unit within eukaryotic chromatin. It is composed of double-stranded DNA wrapped around an octamer of histone proteins. In eukaryotes, it is an octamer of histone proteins.
DNA Supercoiling
The process of the DNA double helix winding upon itself, the formation of additional coils in DNA due to twisting forces.
Mechanisms that make the bacterial chromosome more compact include… (To fit within a bacterial cell, the chromosome must be compacted about 1000-fold.)
DNA supercoiling
The formation of microdomains and macrodomains
Negative supercoiling promotes strand separation. Why is strand separation beneficial? (In living bacteria, the DNA is negatively supercoiled.)
Because it enhances genetic activities such as replication and transcription that require the DNA strands to be separated.
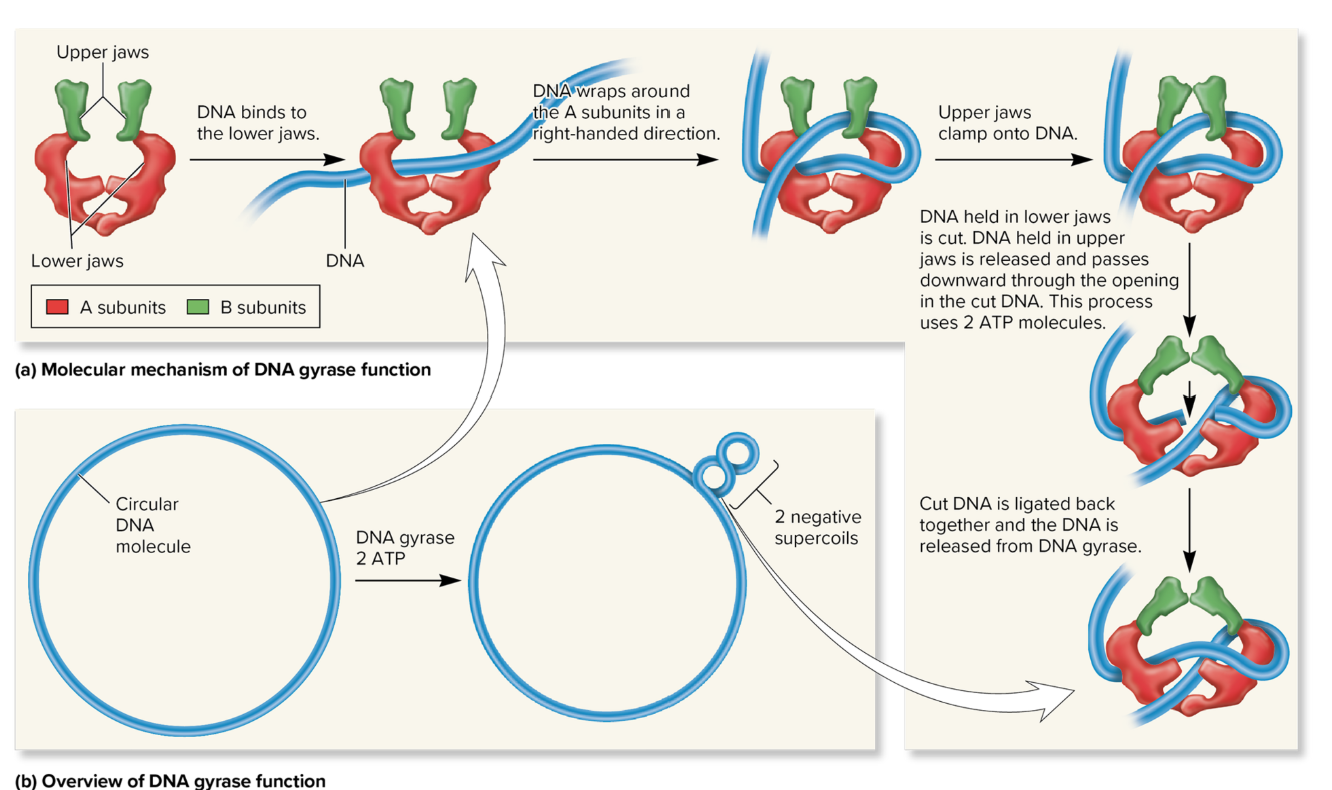
DNA Gyrase
an enzyme that introduces negative supercoils into DNA using energy from ATP and that can also relax positive supercoils when they occur. Also known as topoisomerase II.
DNA gyrase
promotes negative supercoiling.
relaxes positive supercoils.
cuts DNA strands as part of its function.
does all of the above
does all of the above
Negative supercoiling can enhance RNA transcription and DNA replication because it
allows the binding of proteins in the major groove.
promotes DNA strand separation.
makes the DNA more compact.
causes all of the above.
promotes DNA strand separation
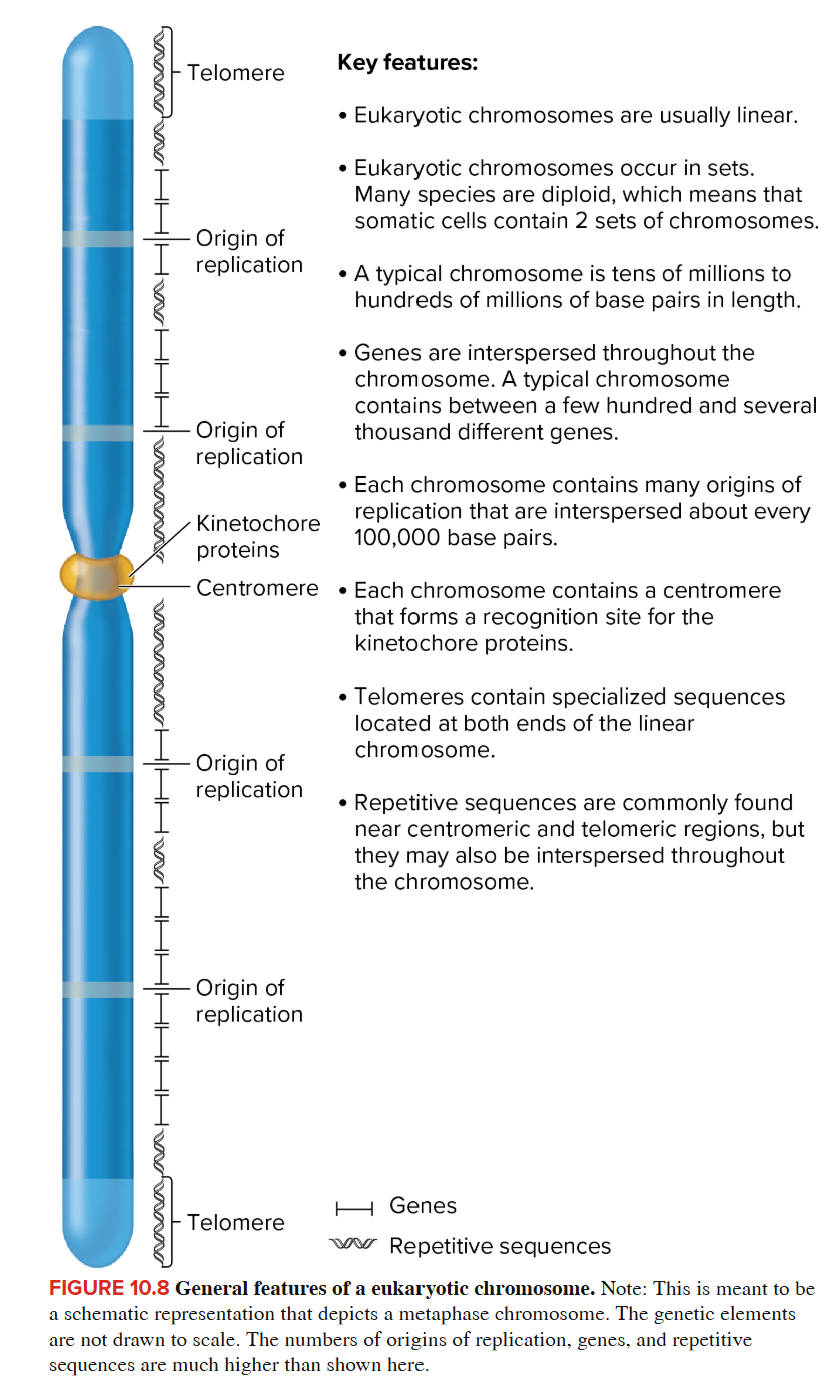
Eukaryotic Chromosomes
Contains a long, linear DNA molecule
Occur in sets, many species are diploid, which means that somatic cells contain 2 sets of chromosomes. Humans have 46 chromosomes, 2 sets of 23 chromosomes each (one set from mom, one set from dad).
Tens of millions/hundreds of millions of base pairs in length
Contains multiple origins of replication
Contains a centromere that forms a recognition site for kinetochore proteins
Has telomeres at both ends that contain specialized sequence
Repetitive sequences commonly found near centromeric and telomeric regions.
Kinetochore
A group of proteins that link the centromere to the spindle apparatus during mitosis and meiosis, ensuring the proper segregation of the chromosomes to each daughter cell.
Telomeres
Specialized repeated sequences of DNA found at the ends of eukaryotic chromosomes.
prevent chromosomal rearrangements such as translocations.
prevent chromosome shortening in two ways. 1. protect chromosomes from digestion via enzymes called exonucleases that recognize the ends of DNA. 2. a specialized form of DNA replication occurs at the telomeres so that eukaryotic chromosomes do not become shortened with each round of DNA replication
Topoisomers
DNA molecules that are structurally the same but differ with regards to supercoiling
Topoisomerase I
One of the structures that controls supercoiling (other being DNA gyrase), an enzyme that alters the degree of supercoiling in DNA by relaxing negative supercoils.
Compacting Eukaryotic Chromosome
The compaction of linear DNA in eukaryotic chromosomes
involves interactions between DNA and several different proteins
Eukaryotic vs Bacterial DNA
The total amount of DNA in eukaryotic species is typically much greater than that in bacterial cells - contains many more genes.
Eukaryotic genomes vary substantially in size but this variation is not related to the complexity of the species because
The difference in the size of the genome is not because of extra
genes. Rather, the accumulation of repetitive DNA sequences! (not because there are more genes, so its more complex, but bigger because there are more repetitive DNA sequences that do not code proteins)
Sequence complexity
Refers to the number of times a particular base sequence appears in the genome
3 main types of repetitive sequences
Unique or non-repetitive
Moderately repetitive
Highly repetitive
Type of repetitive sequence: Highly repetitive
Found tens of thousands to millions of times, each copy is relatively short (a few nucleotides to several hundred in length)
Type of repetitive sequence: Moderately repetitive
Found a few hundred to several thousand times, genes for rRNA and histones, sequences that regulate gene expression and translation.
Type of repetitive sequence: Unique or non-repetitive
Found once of few times in genome
Some sequences are interspersed throughout the genome
LINEs (Long interspersed elements)
• Usually 1,000 to 10,000 bp long
• Occur in 20,000 to 1,000,000 copies per genome
• Almost 17% of the human genome
SINEs (Short interspersed elements)
• Less than 500 bp in length
• Example: Alu sequence present in about 1,000,000 copies in
human genome (10% of the genome)
Tandem arrays
A region in DNA in which a very short nucleotide sequence is repeated many times in a row. Also known as a tandem repeat.
Example: ATAT and AATATAT sequences in Drosophila
• These are commonly found in the centromeric regions
Which of the following is/are moderately repetitive sequences?
How long would a single set of human chromosomes be?
If stretched end to end, a single set of human chromosomes would
be over 1 meter long!
• Yet the cell’s nucleus is only 2 to 4 μm in diameter
• Therefore, the DNA must be tightly compacted to fit within the
nucleus
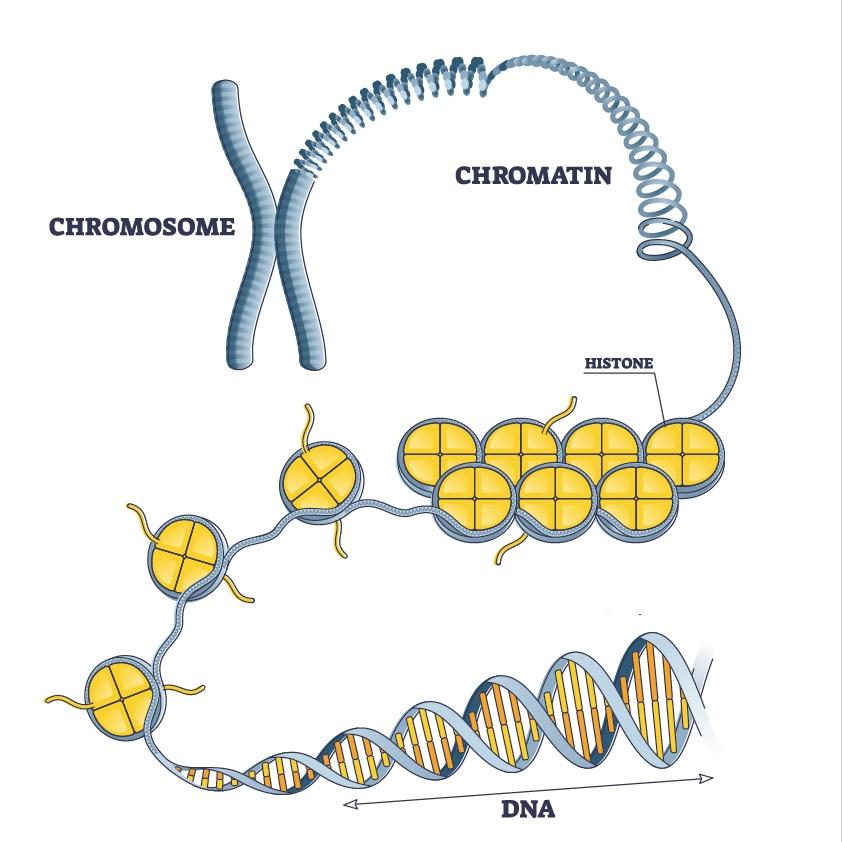
Chromatin
The complex of DNA, and proteins that forms chromosomes within the nucleus of eukaryotic cells.
Proteins bound to DNA are subject to change during the life of
the cell, these changes affect the degree of chromatin compaction
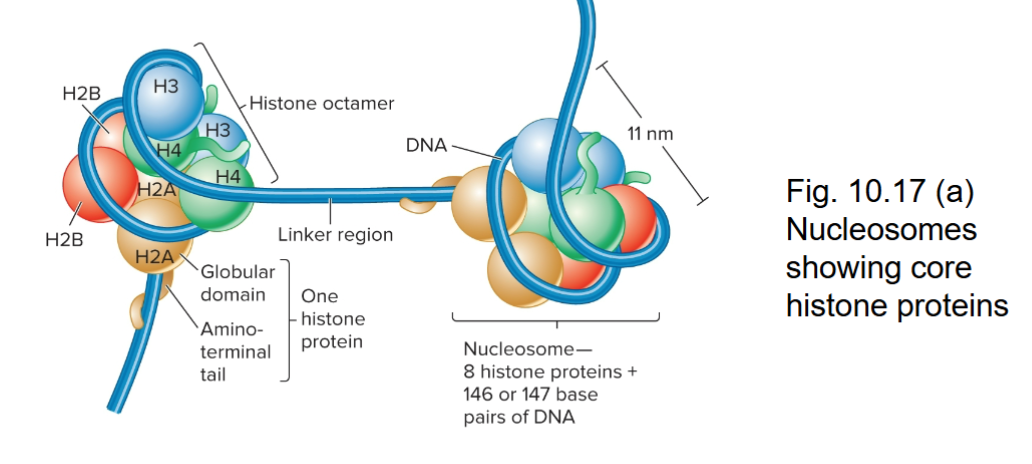
Histone Octamer
Composed of two copies each of four different histone proteins
146 base pairs of DNA wrap almost two times (1.65 times) around a ball made of histone proteins, forming a compact structure called a nucleosome. This wrapping introduces a small negative twist that helps the DNA pack tightly but still be accessible for cell processes.
If you assume that the average length of a DNA linker region is 54
bp, approximately how many nucleosomes can be found in the
haploid genome, which contains 3 billion (3,000,000,000) bp?
Note: Nucleosome:~146 bp of DNA wrapped around a histone
octamer.
Linker (54bp) +Nucleosome (146bp) = 200bp
3,000,000,000/200bp = 15,000,000
Histone Proteins
Contain many positively charged amino acids
Bind to the negatively charged phosphates along the DNA backbone
Five types of Histones
H2A, H2B, H3, and H4 are the core histones, H1 is the linker histone
Two of each core histones make up the octamer
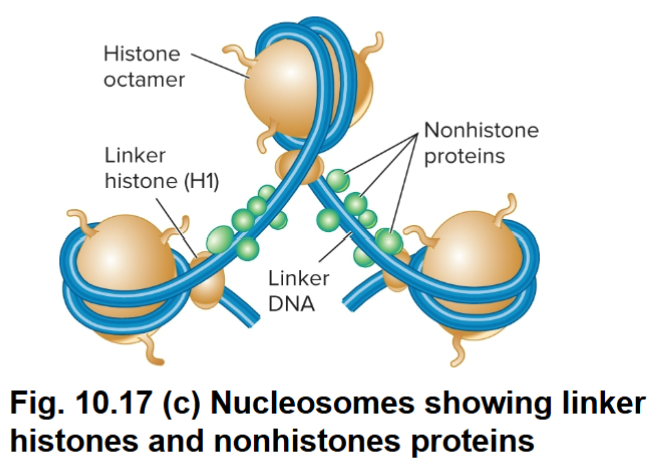
H1: Linker Histone
Helps to organize adjacent nucleosomes
Linker histone binds to DNA in the linker region
Linker region is less tightly bound to DNA than core histones
Further compaction of the Eukaryotic Chromosome (first were wrapping of DNA within nucleosomes and the interaction of nucleosomes to form a zigzag arrangement)
Chromatin can be further compacted by folding segments of nucleosomes into loops, or loop domains.
What proteins play a role in loop formation, according to the loop extrusion model?
SMC proteins (structural maintenance of chromosomes)
CCCTC binding factor (CTCF)
Loop extrusion model
A mechanism for loop formation in eukaryotes in which SMC proteins promote the formation of a loop, and the bottom of the loop is held in place by SMC proteins.
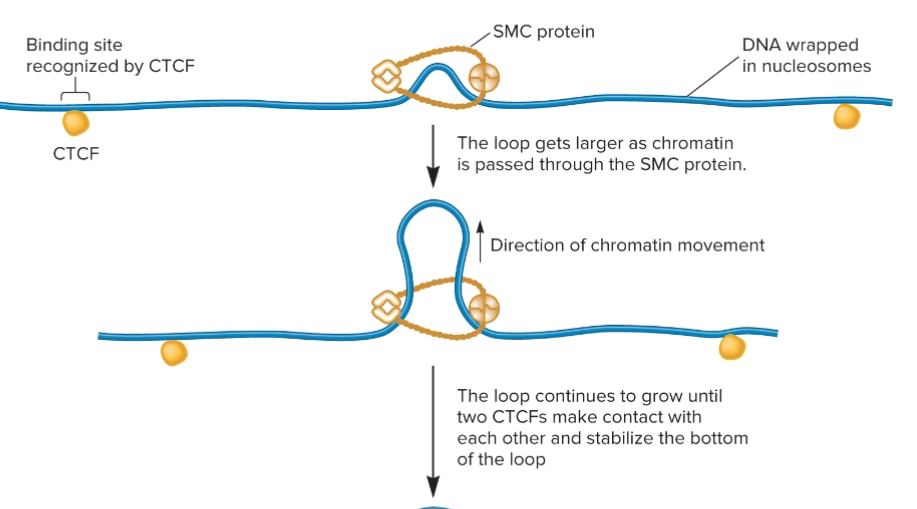
SMC proteins
Forms a dimer that can wrap itself around two DNA segments and promote the formation of a loop
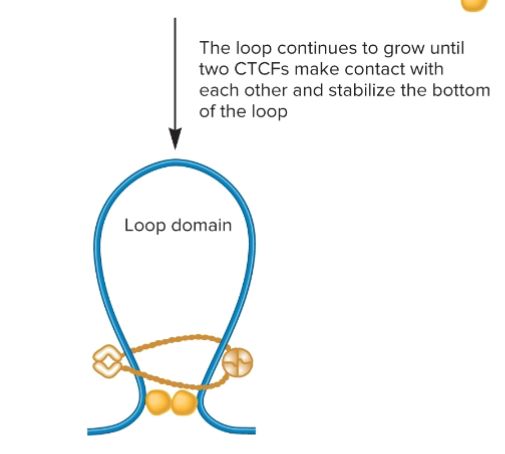
CTCFs proteins
In a loop extrusion model, after a loop has formed due to the action of SMC proteins, two different CTCFs bind to the DNA and then bind to each other to stabilize the loop!
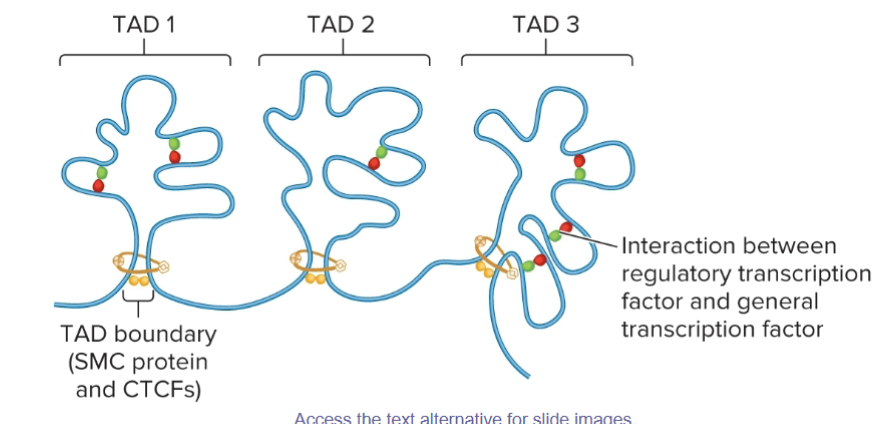
Topologically Associated Domains
a chromatin region in which segments of DNA within each TAD are more likely to interact with each other than they are with segments in other neighboring TADs
Approximately 100 kb to 1 Mb in length.
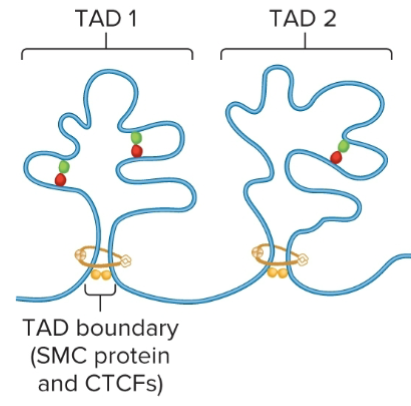
TAD boundaries
SMC Proteins and CTCFs
Act as insulators, preventing interactions between different TADs
Promote interactions within a TAD
Heterochromatin
During interphase, highly compacted regions of chromosomes in which the DNA is usually transcriptionally inactive.
Loop domains compacted even further
Euchromatin
Less compacted regions of chromosomes, where DNA is transcriptionally active.
Loop domains are less compacted
Type 1 of Heterochromatin: Constitutive heterochromatin
Regions that are always heterochromatic
• Permanently inactive with regard to transcription
• Usually contain highly repetitive sequences
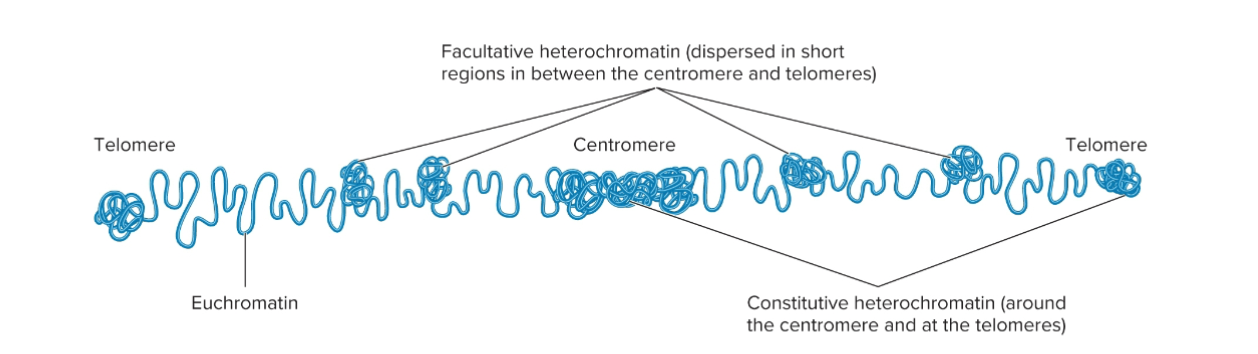
Type 2 of Heterochromatin: Facultative heterochromatin
Regions that can interconvert between euchromatin and
heterochromatin
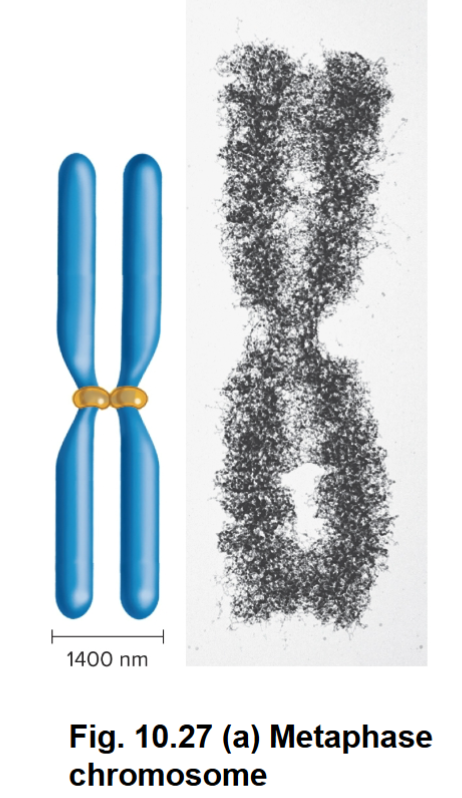
Structure of Eukaryotic Chromosomes During Cell Division
As cells enter M phase, the level of compaction changes dramatically
• Sister chromatids are entirely condensed
• Two parallel chromatids have an overall diameter of 1,400 nm and are much shorter than in interphase
Metaphase Chromosomes
The highly compacted form of a eukaryotic chromosome that is observed during metaphase of mitosis.
Undergo little gene transcription
Chromatin within metaphase chromosomes is organized along a scaffold (scaffold contains SMC proteins)