BIO 2B03: Modules 1-4
1/221
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
222 Terms
What are the 7 main functions of proteins
Structural component of cells
Sensors for environmental changes
Molecular motors (on/off switch)
Gene regulation
Enzymes (catalysts for chemical reactions)
Signalling molecules between cells
Organelle identity and fucntion
What are the 4 main components of an amino acid?
Carboxyl group (carboxylic acid without the H on OH)
Amino group (NH3+)
Hydrogen side chain
Variable R group
What component of an amino acid affects the protein structure and function? How does this happen?
The variable R group
Interactions of the side chains determine how the protein folds
What does it mean to say a molecule is soluble? What makes a molecule soluble?
Physical property that the molecule can impermanently bond to water through hydrogen bonding, and soluble proteins will typically carry hydrophilic amino acids on their external surface
Molecule that can form H bonds with water = soluble
Cannot form H bonds = insoluble
What is said about the charges for hydrophilic vs hydrophobic molecules?
Hydrophilic: charge polarized (delta +, delta -)
Hydrophobic: not charge polarized
What types of molecules are hydrophobic?
Saturated hydrocarbons (oils, fats, alkanes)
What are the 2 groups of hydrophobic amino acids?
They have non-polar side chains:
Aromatics (phenylalanine, tyrosine, tryptophan)
Aliphatics/Hydrocarbons (alanine, valine, isoleucine, leucine, methionine)
What characteristics do hydrophilic amino acids typically have? (3)
Charged/ionized at physiological pH (7)
Has an -OH at one end (O-) OR
Has an -NH2 at one end (NH3+)
What are the positively charged amino acids?
Lysine and Arginine (has the NH3+ groups)
What are the negatively charged amino acids?
Aspartic acid and glutamic acid (has the O- groups)
Which amino acids are polar uncharged?
Serine and Threonine
have polar OH groups but are uncharged at neutral pH
Asparagine Glutamine
have polar amide groups but are uncharged at neutral pH
Which amino acid can form disulfide bridges? How does it happen?
Benefits?
Cysteine
covalent bonds between sulfur atoms and OTHER cysteine molecules
Strength, decreased flexibility
Which amino acid can tuck in for tight folding? What does this allow for?
Glycine with the single H variable side chain
Allows for bends in the polypeptide, which makes it more flexible
Which amino acid forces a kink in the peptide chain? How does this happen?
Proline
The R-group covalently bonds to the N in the amino group, which forms a kink/bend
Which amino acid can shift charge depending on pH of environment? Benefits?
Histidine
Nitrogen shifts from neutral to + charge when pH gets lower (~5)
Can facilitate reactions or stabilize other molecules, and maintains structure under pH changes
How are peptide bonds formed?
Condensation reaction between amino group and carboxyl group of separate amino acids
What is the meaning of N-terminus and C-terminus? Where are they located?
N-terminus: amino end on left
C-terminus: carboxyl end on right (where the next amino acid is added during translation)
T or F: proteins have a single stable structure
F, not always, sometimes they have a collection of related structures they switch between
What is a periodically ordered structure?
When polypeptides spontaneously assume a random coiled structure
What is a statistical coil?
A structure that a protein spends most of its time in (may be stabilized by interactions within the polypeptide or between other proteins/molecules
T or F: interactions that maintain protein shape are typically non-covalent
T
What is the hydrophobic effect?
Nonpolar hydrophobic molecules combine to minimize surface area exposed to hydrophilic aqueous environments (eg oil droplet in water)
What is the secondary structure of a protein? What are they defined by?
Periodic folding into distinct, conserved geometric arrangements (a-helix, b-sheet)
Defined by: patterns of H-bond formation that occur between non-variable side chains of amino and carboxyl groups (forms INDEPENDENTLY of r-groups)
In an alpha helix and beta sheet, what is the structure behind the shape?
H-bonding between the O in carboxyl group and H in amino group
4 POSITIONS AWAY for a-helix
between 2 parallel aligned polypeptide stretches
T or F: b-sheets are always formed between 2 separate polypeptides
F, can be formed within a single polypeptide
What is a beta-turn?
involves 3-4 amino acids, and is found connecting the strands of a b-sheet (basically the corner)
What is a motif?
Particular combinations of secondary structures, 3D
What are the 4 main motifs and their structres?
Coiled-coil motif (2 a-helices wrapped around each other)
Hydrophobic residues form a continuous hydrophobic surface, wrapping around each helix causing each helix to wrap around each other
Zinc-Finger motif (a-helix and 2 b-strands)
Held in position by interaction of precisely places Cys or His residues with a zinc atom
B-barrel motif (4-10 antiparallel beta strands form a sheet & channel)
b-sheet forms a barrel when last b-strand hydrogen bonds with the first strand
Helix-loop-helix motif (2 a-helices joined by a loop region)
Calcium facilitates the shape through non-covalent interactions between itself and Asp or Glu
What motif is a leucine zipper representative of?
Coiled-coil
What is the tertiary structure of a protein? What is a fundamental unit of it?
Overall conformation of a single polypeptide
Domain: substructure produced by any part of the chain that can fold independently into a stable, compact structure
What are the 2 types of protein domains?
Functional Domain (regions of a protein that perform a certain activity)
eg DNA binding, protein-protein interaction
Structural Domain (regions of a protein that form compact, independent globular domains)
eg acidic domain, proline-rich
Quaternary Structure of a protein
Assembly of a multimeric protein (multiple separate proteins
What are the following terms?
a) Dimer
b) Trimer
c) Homodimer
d) Heterodimer
a) 2 polypeptide or subunits
b) 3 polypeptides
c) 2 identical polypeptides
d) 2 different polypeptides
What is an intrinsically unstructured protein?
Proteins that lack a tertiary structure by themselves, but when paired with another IUP forms a stable functional protein
Can require a substrate or another unstructured protein to evolve out of the unstructured state
T or F: stabilization of an unstructured protein happens as an SN1 reaction
F, the recognition of a substrate and stabilization happen simultaneously which makes it an SN2 reaction
What is an example of an intrinsically unstructured protein becoming stable?
Zinc finger complex recognizes DNA and becomes a stable complex
What post-translational modification protects against protease degradation
Acetylation
What post-translational modification alters gene expression by weakening association between histone proteins and DNA
Methylation
What do phosphatases do? What does the opposite of it?
Removes a phosphate group
Kinases transfer phosphate groups TO the amino acid
What are the post-translational modifications hydroxylation and carboxylation?
Hydroxylation: addition of OH group
Carboxylation: addition of COO- group (adds a negative charge which can facilitate ionic bonding)
What post-translational modification protects against proteolysis, and helps with proper protein folding? Where does it occur?
Glycosylation, addition of carbohydrates
Golgi apparatus
What post-translational modification anchors proteins to membranes?
Lipidation
What is the native state of a protein
Most thermodynamically stable conformation of a protein
What did Christian Anfisen do and test in his protein experiment?
He denatured proteins with high concentrations of urea which broke H-bonds and disrupted hydrophobic interactions
Then dialyzed (removed denaturants) the protein
The protein ended up refolding into the same conformation as before, showing that protein reassembling is possible
Why does sickle-cell anemia occur?
A single amino acid change affects the protein structure by changing a charged amino acid to a hydrophobic residue, causing the final RBC to be sickle-shaped
All proteins are linear _________ of amino acids, individually known as __________
polymers
monomers
3 rules for protein folding:
Spontaneous
Reversible
unique
What are genetic amyloid-related diseases
single changes in nucleotides that change amino acid sequence, leading to unstable intermediates that can form fibrils (accumulations of misfolded proteins that aggregate together)
What are the hereditary disorders vs prion-based disorders talked about in class?
Hereditary:
cystic fibrosis
alzheimers disease
tay sachs
Prion-based:
Mad cow disease
Creutzfeld-Jacob disease
Fatal familial insomnia
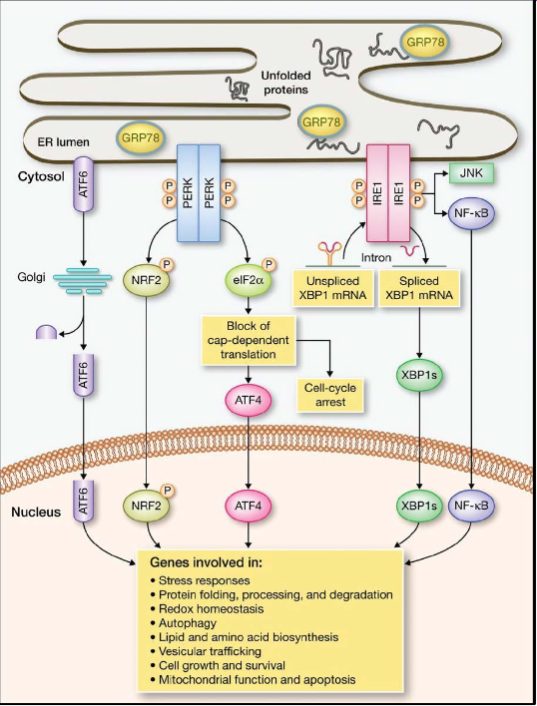
What process do unfolded proteins go through when they accumulate in the ER lumen?
They become bound to GRP78
ER stress sensors are activated (ATF6, PERK and IRE1), inducing a signaling cascade called UPR
UPR down-regulates translation, and activates transcription factors that regulate genes promoting ER homeostasis
During prolonged UPR, apoptosis (cell death) inducing genes are up-regulated
T or F, prion diseases are transmissible
T, they are transmissible and neurodegenerative
What are the characteristics of prion diseases
Lengthy incubation period with irregular immune responses
Spontaneous, heritable, infectious
What is a prion?
abnormal form of a normal protein, attacks other proteins to make them shaped like itself
When is a normal protein more susceptible to prion conversion?
In the transient unfolded state
What are some differences between the PrPc and PrPsc proteins?
PrPc: normal form
3% b-sheet
Monomeric
Soluble
Protease sensitive
PrPsc: disease-associated isoform
43% beta sheet
aggregated (larger association of proteins)
insoluble
protease resistant
What is PRNP
human prion protein gene
missense, insertion, deletion and nonsense mutations cause prion disease in these proteins due to conformational changes that results
What is the path of PrPc cellular trafficking?
PrPc proteins are synthesized, folded and glycosylated in the ER
GPI anchor attaches to the c-terminus as a post-translational modification
2 fatty acids in the GPI anchor the protein to the cell membrane
Conversion of PrPc into PrPsc happens when they come into close proximity which can happen on the surface of the cell, in endosomes or lysosomes
Where does PrPsc accumulate and what does it do?
Accumulates in recycling endosomes and impairs golgi trafficking of membrane proteins such as PrPc
What was significant about the “surgical exposure to brain-eating protein” article
The young epileptics were exposed to CJD contaminated electrodes after they were used 2 years earlier on a patient with CJD. They were exposed to the prions of the previous patient which then infected their proteins and they died
What is an example of a good prion aggregate?
Mod5, a yeast protein has evolved prion aggregates that resist fungicide, by fast on-demand conversion in stressful situations. The resistance to fungus is beneficial
What are the 2 main purposes of chaperones
Prevent inappropriate interactions between amino acids
Increase efficiency of protein folding
Difference between chaperones and chaperonins
Chaperones - monomeric
Chaperonins - multimeric, isolation chamber
What are heat-shock proteins? (HSP)
What do they do?
An example of a molecular chaperone, in mitochondria
Bind to hydrophobic R-groups and prevent the nascent polypeptide from associating with other proteins or from folding prematurely, and aggregating with other hydrophobic residues
When is HSP upregulated and why?
In high-temp environments, because most proteins denature so more HSP is created to refold proteins under these high-stress conditions
Can also be upregulated in cold shock, dessication (removal of moisture), anoxia (loss of oxygen), and exposure to heavy metals and ethanol
What steps are involved in the process of HSP doing its job?
Hydrophobic residues on HSP-70 allow it to rapidly bind to the hydrophobic patches on unfolded proteins
ATP→ADP hydrolysis changes the conformation of HSP-70, which changes the shape of the target protein allowing it to fold properly
ATP hydrolysis is stimulated by DnaJ or HSP-40, and ADP is released from HSP-70 which is assisted by the nucleotide exchange factor GrpE/BAG1
A new ATP fills the nucleotide binding domain, the folded protein is released and HSP-70 is ready for the next protein
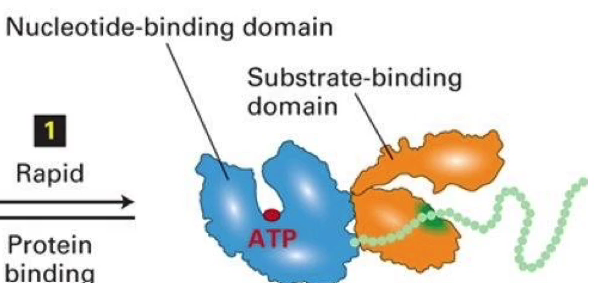
What are the 2 domains of HSP? What do they do?
Nucleotide-binding domain: binds to ATP
Substrate binding domain: unfolded protein
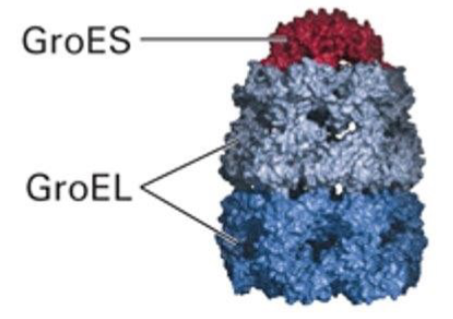
What is GroEL? What does is consist of?
A chaperonin found in bacteria and chloroplasts
Consists of a GroES cap, and 2 GroEL subunits which form the walls and 2 independent folding chambers that CANNOT be used simultaneously
What are the steps involved with GroEL performing its function?
What happens if it doesn’t work?
Bottom chamber releases GroES cap & ADP, while top chamber binds to ATP and a new peptide, as it enters the chamber
New GroES cap binds to the top of GroEL to isolate the substrate peptide
A conformational change enlarges the chamber and allows the peptide room to fold
ATP hydrolysis allows the GroES cap to come off and the folded protein to diffuse out
If the protein is still not folded, it may undergo the process again, but will use the other chamber
What conformational changes occur when GroES binds to GroEL?
GroEL (chamber) shifts to a relaxed conformation and the chamber enlarges to allow room for folding and easy exit of the protein
In bacterial GroEL, what kind of protein subunits make up the structure?
7 HSP-60 subunits form the outside of the protein, and each one binds to an ATP molecule
Each HSP-60 subunit has:
apical domain (top)
Intermediate domain (middle)
Equatorial domain (bottom)
Which protein is in relaxed conformation?
The one with the cap (right)
What is ubiquitin? What is it for?
A small protein with a unique conformation, that is used to tag proteins through ubiquitinylation for degradation
What is the function of a proteasome?
Degrade tagged proteins by cleaving them into small peptide sequences
What does ubiquitinylation require?
3 enzyme system:
E1 - ubiquitin activating enzyme (recognizes Ub and picks it up)
E2 - ubiquitin conjugating enzyme (facilitates attachment of ubiquitin to target protein)
E3 - ubiquitin ligase (recognizes the specific target for degradation & attaches ubiquitin to it)
What are the steps of ubiquitinylation?
Ubiquitin is activated by linkage to E1, which requires energy from ATP hydrolysis
Activated ubiquitin is transferred to Cys on E2
E3 recognizes substrate, and interacts with E2 who transfers the ubiquitin to lysine side chain of the target substrate
More and more ubiquitins are added, since 1 tag is not enough to facilitate degradation
What is the structure of a proteasome?
Wall of many identical subunits create a hollow interior chamber w/ caps at ends
Center contains proteolytic enzymes that break down any protein within the core
Caps have narrow openings where unfolded polypeptides can be threaded through
Has 2 19S caps and one 20S core
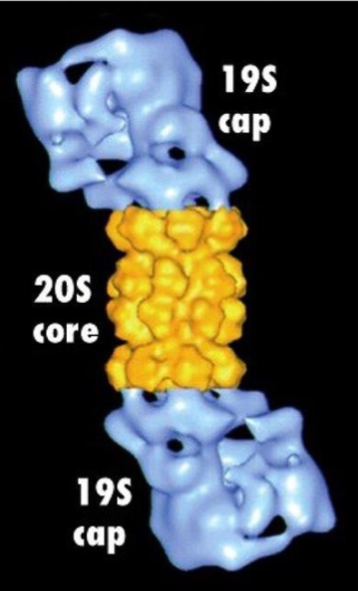
What happens to ubiquitin as the tagged protein is degraded?
Cap of proteasome recognizes polyubiquitin tag
Ubiquitins are removed prior to proteasome entry so they can be reused
What is a ligand?
aka substrate, a molecule that is bound by a protein
What must ligand bonding demonstrate?
Specificity: ability to bind to one or a small number of molecules and not others
High affinity: strength of binding between protein and ligand (strong means they are associated for a long time, weak means the immediately fall apart)
What is molecular complementarity
When the SHAPES of molecules fit well enough, such that favourable, NON-COVALENT interactions conform because all intermolecular forces are contributing (LDF)
(shapes fit well = strong, shapes prevent closeness = weak)
Where are the amino acids facing on a protein in the ligand-binding pocket?
Towards the binding site, will be a specific shape and have specific amino acids for optimal binding affinity
What does a high Keq and low Keq mean for ligase-protein affinity? What about Kd?
Keq = [LP]/[L][P] (L + P → LP)
Kd = [L][P]/[LP] (LP → L + P)
High Keq → high affinity because reaction pushes together
Low Keq → low affinity because reaction pushes to stay apart
High Kd → low affinity because reaction pushes apart
Low Kd → high affinity because reaction pushes together
What effect do enzymes have on a reaction?
They increase the rate of reaction by lowering the activation energy for the transition state
What are the functional regions of an enzyme’s active site?
Binding site/pocket (determines specificity)
Catalytic site (promotes reaction)
What is Vmax regarding enzyme kinetics? what about Km?
Vmax: maximal velocity of a reaction at saturating substrate concentrations
Km: (Michaelis constant) measure of the affinity of an enzyme for the substrate, halfway to Vmax on a graph
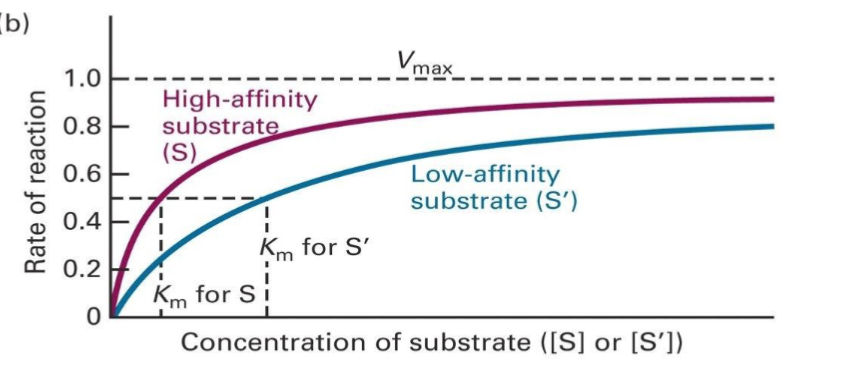
T or F: Vmax of low and high affinity substrates are around the same
T, it just takes a longer, and a higher concentration of low affinity substrate to get to Vmax, since the substrate and enzyme are more frequently dissociating
What does decreasing the concentration of the same enzyme do to the reaction?
Decreases reaction rate, since there is less of the enzyme available
What is PKA’s structure, and how does it relate to its function?
Has nucleotide binding pocket (for ATP)
Glycine lid
Peptide binding pocket
BOTH BINDING DOMAINS ARE KNOWN AS KINASE CORE
A conformational change occurs once the substrates bind. The nucleotide binding domain will undergo close contact between ATP and target peptide to easily phosphorylate it
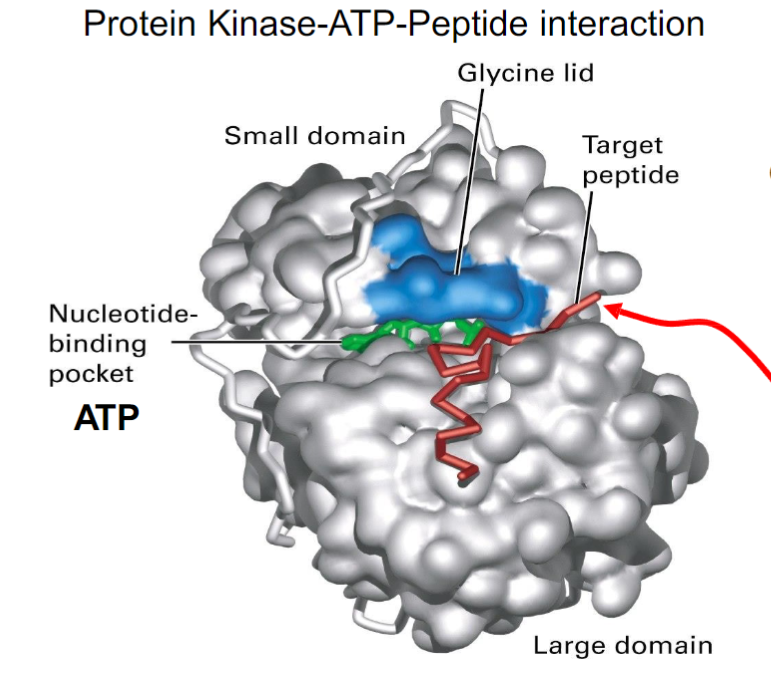
What is the open conformation vs closed conformation of PKA?
Open:
substrates bind, due to binding sites being exposed
ADP and phosphorylated peptide are released after
Closed:
Glycine-rich lid traps substrates
Phosphorylation of the target peptide occurs
How do substrates detach when PKA reopens?
ADP and the phosphorylated protein can simply fall off, since their affinity for the enzyme binding sites is no longer high
What 5 general mechanisms regulate protein function?
Allosteric regulation
Signal-induced regulation of protein levels
Covalent modification
Proteolytic cleavage
Enzyme complexes
What do allosteric modulators do?
small molecules that bind to sites other than the active site of a protein, to induce a conformational change and modify the protein function
Positive modulators: change protein shape to increase activity
Negative modulators: decreases activity
What are the active vs inactive states of an enzyme also known as regarding allosteric regulation?
The tertiary vs quaternary structure of the enzyme
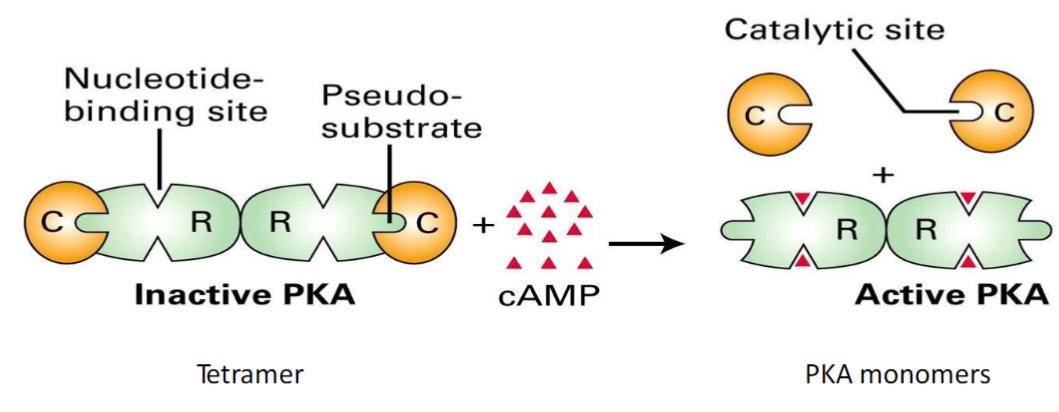
What is cAMP considered in the reaction?
The positive modulator
It binds to the regulatory subunits of PKA, causing a conformational change so the pseudo-substrate changes shape to reveal the catalytic site. The reveal of the catalytic site will allow the enzyme to be active = positive modulation
What happens when cAMP is bound to the allosteric modulator site in PKA? What happens when its released?
Bound - the pseudo substrate domain is retracted, and can’t bind to the catalytic domain leaving PKA ACTIVE
Released - conformation of the pseudo substrate site changes to extend outwards so it can interact with the catalytic site, which blocks the binding site and leaves PKA INACTIVE
What affect does CTP have when it binds to Aspartate transcarbomylase? What would CTP be classified as?
When CTP binds to the regulatory subunits (arms), it masks the substrate binding sites and twists the entire complex into a tense conformation, making it inactive
The enzyme becomes active again once CTP detaches
CTP is considered an allosteric inhibitor
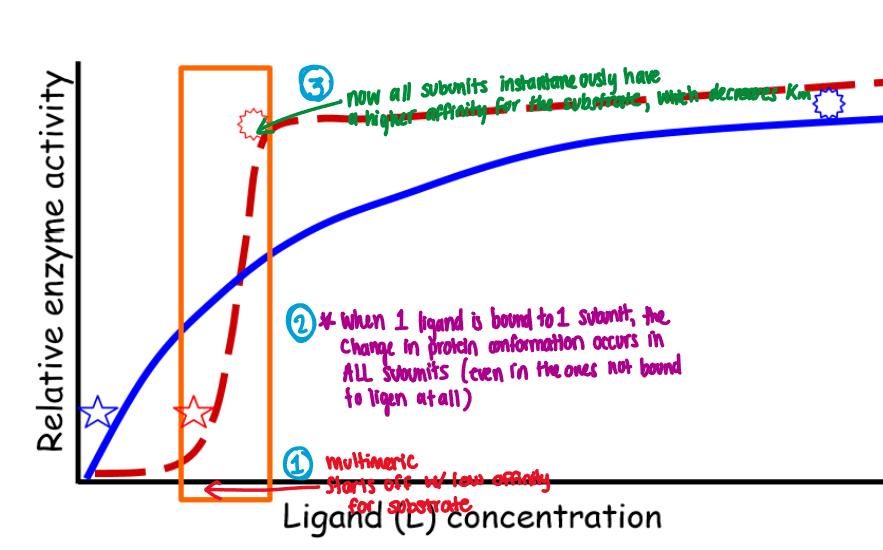
Cooperative allostery
When the binding of one ligand molecule affects the binding of subsequent ligand molecules
Example:
Enzyme starts off with low affinity for substrate
When 1 ligand is bound to 1 subunit, the change in protein conformation occurs in ALL subunits (even ones not bound to ligen)
All subunits instantaneously have a higher affinity for the substrate
What does the binding of oxygen to haemoglobin facilitate?
Moves from inactive state to active state, and changes the conformation of the subunits to have higher affinity for oxygen (oxygen is the substrate AND allosteric activator)
This happens due to different parts of the body needing more oxygen than others (lungs vs tissue)
What is 2,3-BPG with respect to haemoglobin?
An allosteric inhibitor
Present in high concentrations where Oxygen is needed from haemoglobin, because it binds to an effector site on the protein and decreases its affinity for O2
What are the 3 amino acids that can be phosphorylated?
serine
thionin
tryocine
(all have the OH side chain)