Exam 3 DNA repair mechanisms
1/39
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
40 Terms
DNA repair system
maintains integrity of genetic material
counteract genetic damage that would result in genetic diseases and cancer
proofreading and mistmatch repair
DNA polymerase “proofreads”, removes and replaces incorrectly inserted nucleotides
mismatch repair becomes activated if proofreading fails
mismatches are detected, cut, and removed, correct nucleotide inserted by DNA polymerase
exonucleases
cut off the ends of a sequence
endonucleases
cut out mismatches within a sequence
strand discrimination
based on DNA methylation
adenine methylase (enzyme in bacteria) recognizes DNA sequences and adds methyl group to adenine residues
newly synthesized strand of replication remains unmethylated
mismatch repair recognizes unmethylated strand and repairs
DNA polymerase
“proofreads”, removes and replaces incorrectly inserted nucleotides
has a 3’ to 5’ proofreading activity which repairs errors induced by incorrect base incorporation during replication
has exonuclease activity: lowers the rates of errors 100 fold to 10-7 , now 1 mistake in every 10,000,000 bases then 100,000
post replication repair
responds after damaged DNA has escaped repair and has failed complete replication
RecA protein directs recombination exchange with corresponding region on undamaged parental strand (donor DNA)
gap can be filled in by repair synthesis
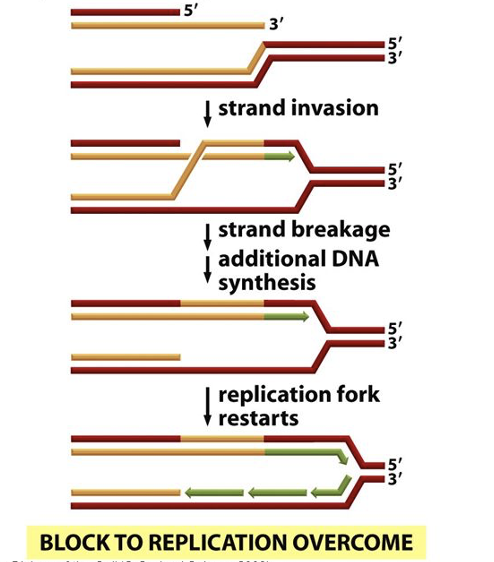
E.coli SOS repair system
last resort to minimize DNA damage
DNA synthesis becomes error-prone; inserts random/incorrect nucleotides in places that normally would stall replication
bacteria induce expression of 20 genes; their products allow replication to move forth
SOS repair can itself become mutagenic; allows cells to survive with DNA damage (cell would otherwise kill itself)
photoreactivation repair (PRE)
seen in E.coli
photoreactivation enzyme cleaves bonds between thymine dimers, reversing effect of UV radiation on DNA
enzyme must absorb photon of light from blue end of the visible light spectrum to cleave dimer
humans and other organisms lack this repair
base and nucleotide excision repair
light-independent DNA repair mechanisms exist in all prokaryotes and eukaryotes and involve excision repair
endonuclease recognizes and cuts distortion/error
DNA polymerase inserts complementary nucleotides in missing gap
DNA ligase seals final “nick”
base excision repair (BER)
corrects DNA containing a damaged DNA base
DNA glycosylase recognizes altered base
different glycosylases for each type of nucleotide and have ability to recognize nucleotides that have had their structure altered by a tautomeric shift, deamination, alkylation or other modification
when recognized, an altered base they attach to it and rotate the nucleotide breaking the hydrogen bond that holds it to the base on the complementary strand, nitrogenous base is then removed from sugar backbone molecule
nucleotide excision repair (NER)
repairs bulky lesions that alter/distort double helix
recognizes mispaired bases or dimers as they cause a distortion in the shape of double helix
purine + purine: too wide or pyrimidine + pyrimidine: too narrow
remove around 20 bases on either side of the incorrect base
DNA polymerase and ligase will come in and add nucleotides and seal “nick'“
Xeroderma Pigmentosum (XP)
rare genetic disorder due to defects in nucleotide excision repair (NER) pathways: inability to repair damaged caused by UV radiation, often have freckles on their skin
affected individuals exhibit severe skin abnormalities, skin cancers, developmental and neurological defects
individuals have 2000-fold higher rate of cancer
heterokaryon cells
formed by fusing two cells together to form one in cell culture, one with ability of NER and one not capable (XP cell)
if cell is unable to carry out unscheduled DNA synthesis (normal) the two cells have deficiencies in the same gene
if the cell can carry out unscheduled DNA synthesis when fused, but not when they are individual cells it means that one cell has one deficient gene and the other cell has a different deficient gene
when together the two genes complement one another
double-strand break repair (DSB repair)
extremely dangerous
results in chromosomal rearrangements, cancer, cell death
this repair reattaches strands: two pathways → homologous recombination repair or non homologous end joining
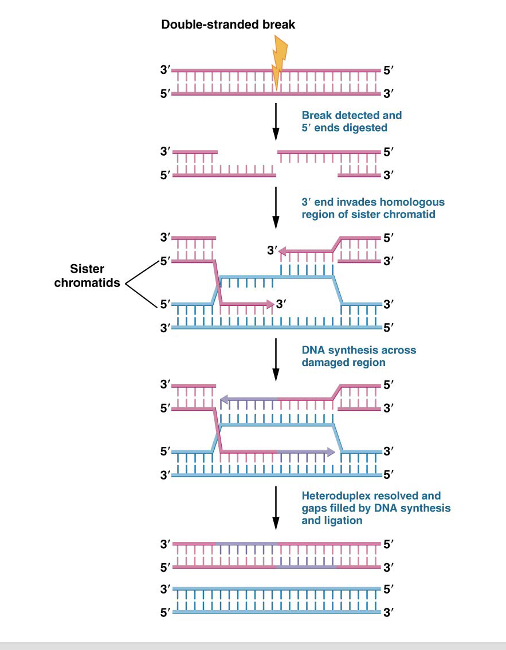
homologous recombination repair pathway
recognizes break, digests 5’ end (done by exonucleases), and leaves 3’ overhang
3’ end aligns with sequence complementary on sister chromatid
sister chromatid is unwound and two strand separate, 3’ ends of the damaged strand base pair with sister chromatid using it as template strand
they extend outward past the point where the break occurred, damaged strands then separate from sister chromatid and base pair with one another
any remaining gaps are filled in by DNA polymerase and DNA ligase while the sister chromatids return to its double stranded configuration
occurs during late S or early G2 phase of cell cycle
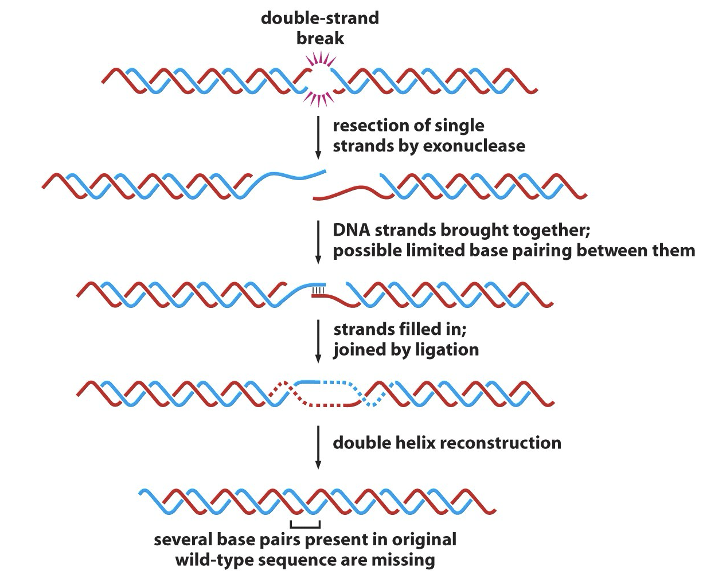
nonhomologous end joining
activated in G1 prior to replication
repairs double-strand breaks
complex of proteins is involved in end joining
may include kinase and BRCA1 (involved in breast cancer)
proteins bind free ends and ligate ends back together
transposable elements (transposons)
“jumping genes” that can move within and between chromosomes
insert themselves into various locations within genome
found in all organisms; precise function is still unknown
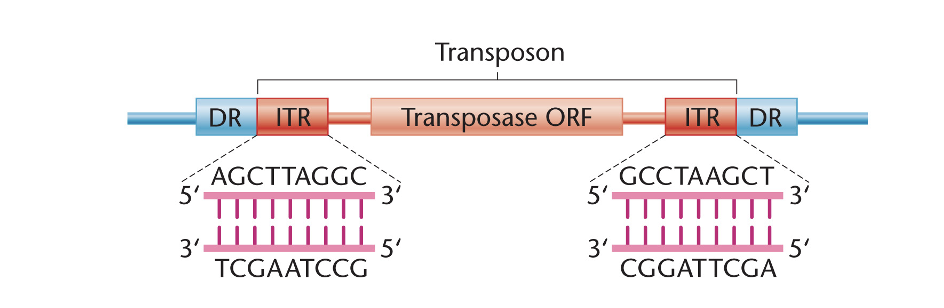
structural features of transposable elements
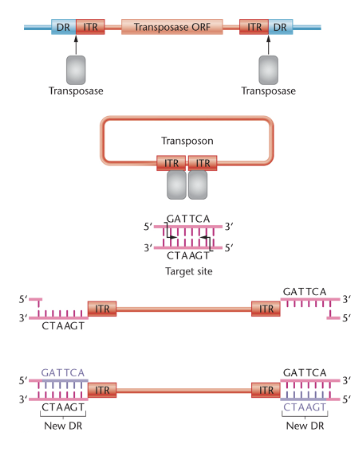
contain an open reading frame (ORF) that encodes enzymes transposase
may also contain ORFs encoding other proteins in addition to transposase
also contain inverted terminal repeats (ITRs) are short DNA sequences that are inverted relative to each other
direct repeats (DRs) flank the DNA transposon in the chromosomal DNA
Steps in DNA transposon transposition
transposase cleaves DNA at ITRs
Transposase makes staggered cuts at target site in chromosomal DNA
transposase inserted transposon at target site
gaps in target site are filled in by DNA polymerase and DNA ligase
Insertion sequences (IS elements)
move from one location to another
cause mutations if inserted into gene/gene-regulatory region
Bacterial transposons
larger than IS elements
can introduce multiple drug resistance to bacterial plasmids; move from plasmids to bacterial chromosomes, spreading multiple drug resistance between strains
Ac-Ds system in Maize
mobile genetic elements in corn plants found using this system
dissociation (Ds) and activator (Ac) are two mutations of transposable elements in corn, mobile controlling elements (transposable elements)
movement of Ds gene is dependent on Ac gene
Barbara McClintock
nobel prize physiology or medicine 1983
found some kernels are white, some are blue and some are speckled because of transposable elements
Effects of Transposable Elements
if a TE inserted into a protein coding region, it can disrupt the protein (null mutation)
if a TE inserts itself into a promoter, it can prevent a gene from being expressed
if a TE that is inserted into a promoter contains a promoter for one of its own genes, it can cause a gene to be expressed when it usually isn’t (or shouldn’t)
types of transposable elements in plants
autonomous elements: can transpose (jump) by themselves, are self contained
nonautonomous: cannot transpose by themselves and require the assistance of an autonomous element to provide the necessary gene products
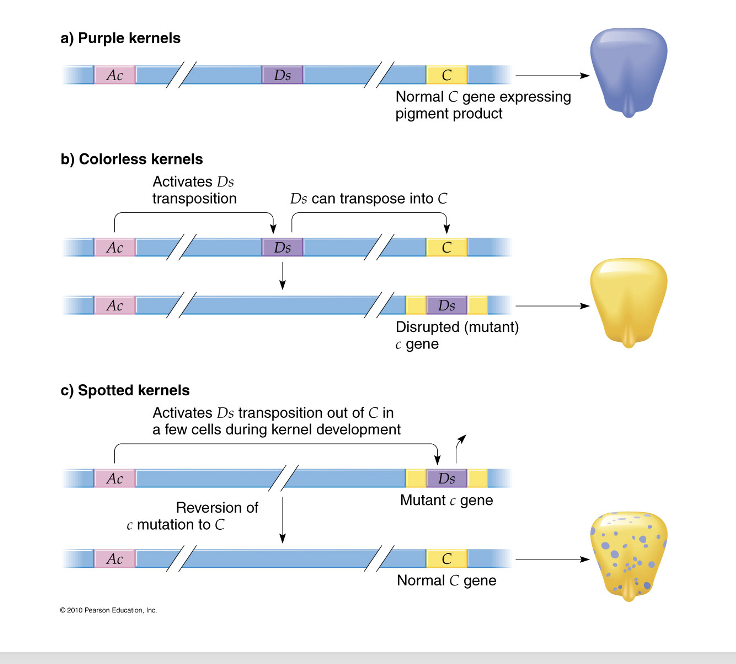
McClintock’s discovery
if a corn plant carries the C gene it produces anthocyanin which makes the kernels purple
some mutants called c mutants are colorless because the anthocyanin is not deposited in the cells
in some cases there is a reversion during development and pigment is deposited in certain cells giving a speckled appearance
if reversion occurs early in development there is more purple tissue in the kernels
hypothesized that the c mutation was occuring when a “mobile controlling element” called Ds for dissociation was inserted into the C gene
Ds requires the help of an activator Ac to insert itself
the Ac activator can also excise the Ds element causing the reversion
Ac and Ds
Ac is an autonomous transposable element: can copy itself, move to a new location and insert itself; can produce the enzyme transposase
Ds is a non autonomous transposable element; it cannot copy itself or move to a new location by itself; requires the assistance of Ac which codes for the enzymes that provide the machinery to copy, move and insert it into a new location; it does not produce transposase and uses the enzyme produced by Ac
Mechanism of Ds/Ac
retrotransposons
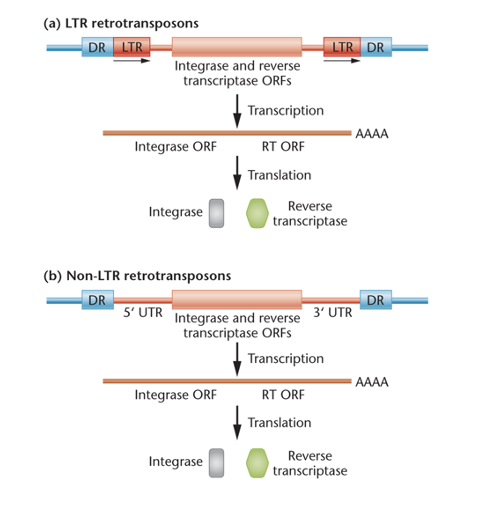
transposable elements that amplify and move within the genome; use RNA as an intermediate
two types: the long terminal repeat (LTR) retrotransposons or the non-LTR retrotransposons
both can be either autonomous or nonautonomous
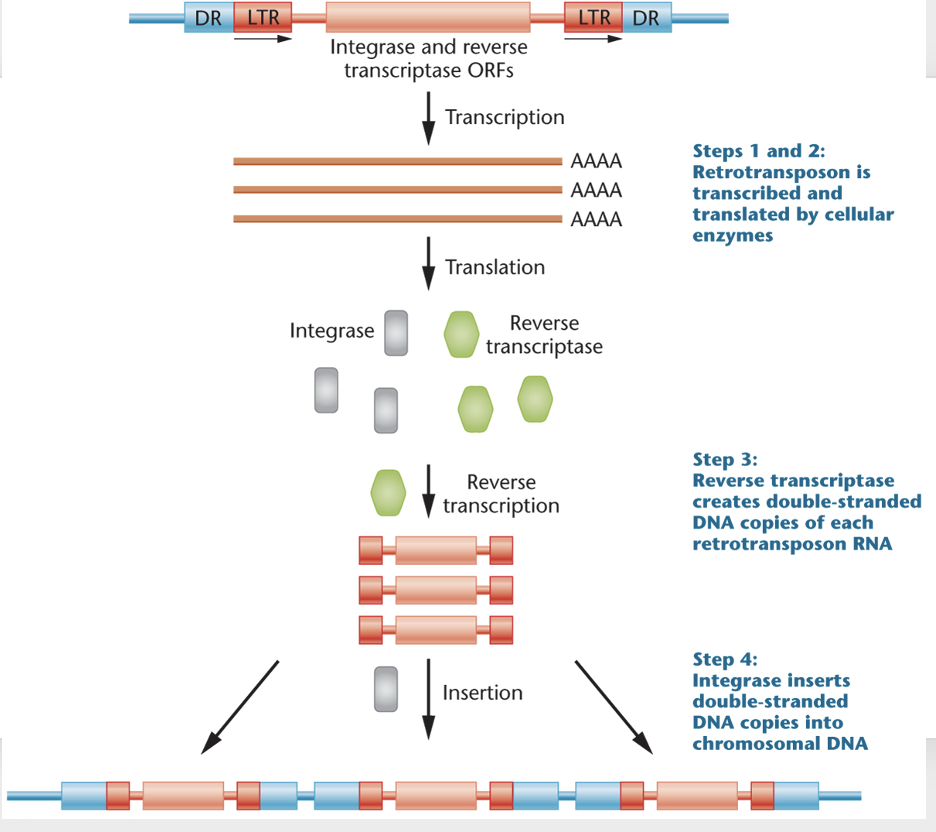
How retrotransposons work
RNA polymerase present in the cell transcribes the retrotransposon
the RNA transcript is translated by the cell’s ribosomes and produces two proteins, reverse transcriptase and integrase
the reverse transcriptase transforms the RNA transcript of the retrotransposon into double stranded DNA
integrase cuts a section of DNA and inserts the DNA version of the retrotransposon into a new location
retrotransposon structure
LTRs contain open reading frames that encode the enzymes integrase and reverse transcriptase (RT)
transcription promoters and polyadenylation sites are located within the 5 and 3 long terminal repeats (LTRs)
non-LTR retrotransposons also encode integrase and reverse transcriptase but are lacking LTRs: transcription promoters and polyadenylation sites are located within 5 and 3 untranslated regions (UTRs)
Steps in retrotransposon transposition
1 +2: Retrotransposons is transcribed and translated by cellular enzymes
reverse transcriptase creates double-stranded DNA copies of each retrotransposon RNA
integrase inserted double-stranded DNA copies into chromosomal DNA
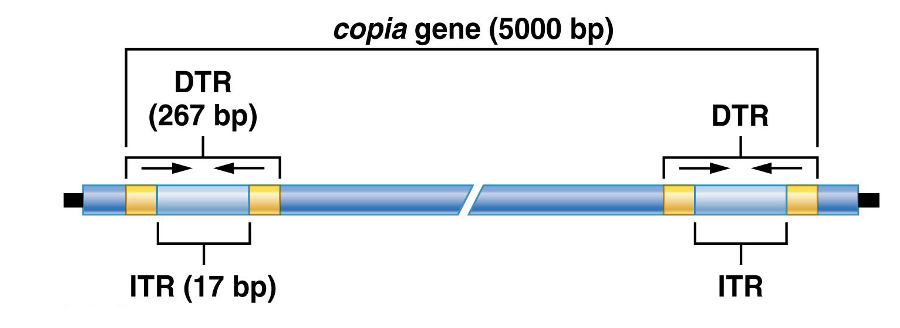
Copia Elements in Drosophila
more than 30 families of TEs found
copia: class of DNA element in Drosophila transcribed into “copious” amounts of RNA
dispersed throughout genome and transposable to different chromosomal locations
(DTR- direct terminal repeat (267 bp long and ITR- inverted terminal repeat 17bp)
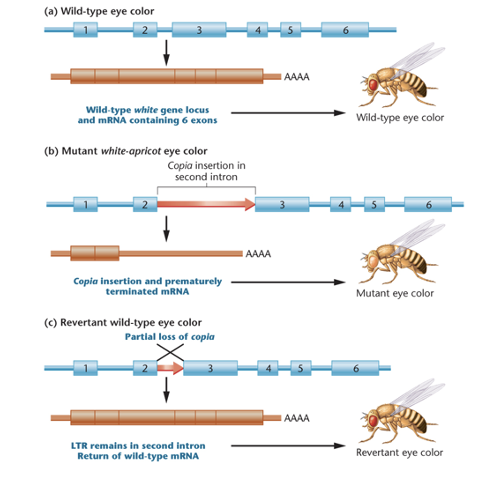
Effects of Copia Elements in Drosophila
a. white gene in wild-type drosophila contains 6 exons, all of which are present in the mRNA
b. the white gene in mutant white-apricot drosophila contains an insertion of copia (red) in the second intron and a prematurely terminated mRNA containing only 2 exons
c. revertant drosophila have lost most of the copia element form the second intron, resulting in wild-type mRNA and wild-type eye color
hybrid dysgenesis
mutation due to high rates of P element transposition in germ line: elevated mutation rates and sterility caused by TEs
P element in drosophila
discovered during study of hybrid dysgenesis
use as tools for genetic analysis: vectors
crosses between lab stocks from the early 1900s and wild caught flies produced extremely high rates of sterility and mutations (hybrid dysgenesis) only occured when wild caught (new) males were crossed to lab stock females from old stocks; reciprocal crosses were not affected
very useful for producing transgenic drosophila: can inject plasmids with P elements and a specific gene into an egg and it will insert the gene into the genome of the host cell’s germ line
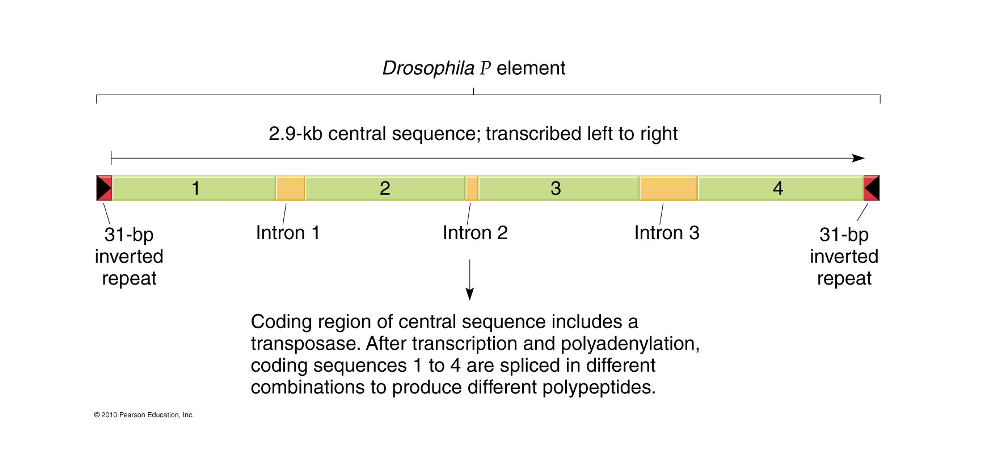
P (new wt strains) and M (old wt) strains
P strains have the P element M strains do not
P strain females produce a protein that suppresses the transposition of the P element, M strain females do not
M strain egg cytoplasm has no protection from transposition leadings to hybrid dysgenesis
P strain eggs have suppressor protein so they are unaffected (also M strain sperm does not contain P elements)
Cross of male/female with or without P element
P male x M Female: sterile because P elements disrupt the genome in eggs
P female x M male: viable because P females produce a suppressor protein that prevents P element transpositions and M males do not have P elements
human transposable elements
half of human genome is composed of TEs
LINEs and SINEs: long interspersed elements and short interspersed elements
0.2% of detectable human mutations may be due to transposable element insertions: transposons may contribute to variability that underlies evolution