U6 mlc genetics megaset
1/152
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
153 Terms
Nucleic Acids
M: nucleotides
P: DNA and RNA
B: phosphodiester bonds
CHONP
Nucleic Acid Functions
information
protein synthesis
regulation
energy
structure
Nucleotide
nitrogen base
5 carbon sugar
phosphate group (1-3)
know how to number sugar carbons
Deoxyribose vs Ribose
Ribose has two OH C-2’ and C-3’
Deoxyribose only OH at C-3’
Nitrogen bases
pyrimidines- single ring (C, T, U)
purines- double ring (A, G)
purines can hydrogen bond to pyrimidines
C-G - 3 H bonds
A-T,U - 2 H bonds
Nucleoside
nitrogen base + sugar
Polymer structure of Nucleic Acids
Outside: Phosphate backbone
Inside: Complementary nitrogen bases
Double Helix
Each strand has a 5’ and a 3’ end
Run in opposite direction (antiparallel)
New nucleotide are added to 3’ end
Synthesized 5’ → 3’ direction!
“We” write 5’ → 3’ direction
“Cells” read 3’ → 5’ direction
Prokaryote vs Eukaryote Genetic Code
Eukaryotes
Linear chromosomes
Nucleus (membrane bound)
Prokaryotes
Circular
Nucleoid (not membrane bound)
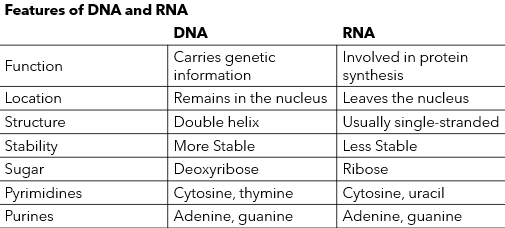
DNA vs RNA (Eukaryotes)
see img
Central Dogma of Genetics
DNA → mRNA→ Protein
Transcription: DNA → mRNA
Translation: mRNA → Protein
RNA breaks down easily → signal is easily regulated
BPQ What is one reason that mRNA made of RNA rather than DNA
Hint: mRNA is a messenger (a signal)
DNA Synthesis vs Transcrip. vs Transla.
Synthesis: DNA → DNA
Transcription: DNA → mRNA
Translation: mRNA → Protein
for cell division
S phase
BPQ
Why does DNA synthesis occur?
In Eukaryotes, when does DNA synthesis occur?
Semi-conservative nature of replication
DNA composed of 2 strands
Each strand provides a template to create a new strand
Each duplicated DNA consists of 2 strands:
1 old strand (template strand)
1 new strand
Replication Steps
Unwind
Unzip
Prime
Copy
Replace
Conjoin
All occurs in close proximity to replication fork
Unwind
Topoisomerase- relaxes DNA supercoiling in front of the replication fork
Unzip
Helicase- breaks hydrogen bonds between two template strands
Single-stranded DNA binding protein
Binds to DNA and prevents reannealing
Prime
DNA replication needs a starting place
Primase- adds an RNA primer from which replication can start rfom
Copy
DNA Polymerase III- creates new DNA strand that is complementary to the template strand
Starts from RNA primer
synthesizes DNA in 5’→3’ direction ONLY
the new strands are made in opposite directions
BPQ
DNA strands are antiparallel, so how will DNA replication look differently for each strand?
Leading and Lagging Strand
bc DNA must be made 5’→3’
Leading Strand
Grows in the 5’ → 3’ direction
Grows continuously
Lagging Strand
Grows in the 3’ → 5’ direction
Grows discontinuously in 5’ → 3’ chunks
Okazaki fragments
Replace
DNA Polymerase I- replaces RNA primers with DNA
Conjoin
DNA Ligase- connects disjointed ends of DNA backbone in the new strands
e.g. between Okazaki Fragments
Proofreading
DNA Polymerase II
Moves slower than DNA polymerase III
Makes less mistakes than DNA polymerase III
Used for proofreading
DNA replication in Prokaryotes vs Eukaryotes
Prokaryote
Chromosomes are circular
Chromosomes are smaller
2 replication forks
1 replication bubble
Eukaryotes
Chromosomes are linear
Chromosomes are larger
Many replication forks
Many replication bubbles
Replisome
The molecular machinery at the replication fork working in unison
Helicase, polymerase, ligase, etc.
Plasmids
Small segments of circular DNA that are separate from the chromosomes and replicate independently
2 major Prokaryote groups
archaebacteria
eubacteria
Central dogma of genetics locations
DNA → mRNA → Protein
Transcription: DNA → mRNA
Nucleus
Translation: mRNA → Protein
Ribosomes (ER or cytoplasm)
RNA
All RNA is created in the nucleus
Messenger RNA (mRNA)
Carries the genetic signal from the nucleus to the ribosomes
Ribosomal RNA (rRNA)
Makes a large part of the physical structure of the ribosome
Transfer RNA (tRNA)
Pulls amino acids into the ribosome to by synthesized as prescribed by the mRNA
Micro RNA (microRNA)
Regulates mRNA translation and breakdown
Post-transcriptional regulation
Transcription
DNA → mRNA
Steps
Initiation
Elongation
Termination
Post-transcriptional* modifications
RNA Polymerase
The multipurpose enzyme that makes pre-mRNA
Multipurpose
DNA synthesis
Unwind – Topoisomerase
Unzip – Helicase
Prime - Primase
Copy – DNA Polymerase III
Replace – DNA Polymerase I
Conjoin – Ligase
Transcription
Unwind – not needed
Unzip – RNA Polymerase
Prime – RNA Polymerase
Copy – RNA Polymerase
Replace – not needed
Conjoin – not needed
in transcription, only opening small segments of DNA → won’t supercoil
BPQ
Why does transcription not need a topoisomerase to unwind the DNA?
Initiation
RNA polymerase binds to a sequence of DNA called the promoter
Promotor contains recognition sites called TATA boxes in Eukaryotes and Archaebacteria
Separates (unzipped) the DNA
Template strand (3’ → 5’)
Coding strand (5’ → 3’)
Elongation
RNA Polymerase reads the template strand (3’ to 5’) and creates pre-mRNA in the 5’ → 3’ direction.
Termination
RNA Polymerase transcribes the terminator signal that causes pre-RNA to separate from the template strand and RNA polymerase
In eukaryotes this terminator transcribes a hairpin loop
Called Rho independent termination
2 types: Rho independent and Rho dependent termination
Rho Independent Termination
Used by Eukaryotes and some prokaryotes
Transcription is terminated by a hairpin loop in the mRNA
Rho Dependent Termination
Used by some prokaryotes
Transcription is terminated by Rho factor
complementary bases will create a hairpin loop (Rho indep. term.)
BPQ
What kind of pattern in the genetic code would create a hairpin loop?
Post-transcriptional Modification
5’ cap and 3’ poly-A tail
Helps protect the mRNA for exonucleases in the cytoplasm
Eukaryotes only
RNA splicing
Removes introns
Pre-mRNA → mature-mRNA
Eukaryotes and Archaebacteria only
mRNA metabolism
RNA is less stable than DNA
mRNA is given a 5’ cap and a 3’ poly A tail to protect it.
Cytoplasm is full of de-capping proteins and exonuclease
Exonucleases – enzyme that removes nucleic acids one at a time
mRNA is intentionally broken down
mRNA can have a half life of a few minutes
RNA Splicing (introns and exons)
Not all the RNA transcribed will be apart of mRNA that leaves the nucleus
Exons -- become part of the final mRNA
Exons “exit” the nucleus
Introns -- are removed
Alternate Splicing
a cellular process in which exons from the same gene are joined in different combinations, leading to different, but related, mRNA transcript and different proteins w/ different structures/functions
One region of DNA can code for multiple proteins by using different exons
Translation
Process of polypeptide chain from mRNA sequence at a ribosome
Prokaryotes
Occurs simultaneously with transcription
No post-transcriptional modifications (in Eubacteria)
Eukaryotes
Must exit the nucleus first
Post-transcriptional modifications
Notice how multiple ribosomes can translate the same mRNA
mRNA doesnt travel far in prokaryotes bc prokary. cells are smaller
also transcrip. and transla. occur simultaneously
BPQ
What are some reason that prokaryotes don’t need a 5’ cap or poly-A tail on their mRNAs?
Codon
4 nucleotides / 20 amino acids
Codon – 3 nucleotides
Start Codon (Methionine)
20 amino acids
Stop Codon
Ribosome
Large subunit
Small Unit
APE sites – sites for tRNA
A- Aminoacyl
P- Peptidyl
E- Exit
Transfer RNA (tRNA)
“transfer” amino acids onto the growing polypeptide
Parts
Anticodon
3 nucleotides that are complementary to a codon**
D loop (do not memorize)
T loop (do not memorize)
Amino Acid attachment site (3’ end)
Only the amino acid that matches the codon may bind here
Wobble Pairing
Sometimes the last nucleotide in an anticodon doesn’t have to match the codon
Translation steps
Initiation – Ribosome assembles around the start codon
Elongation – Amino acids are added to the polypeptide chain
Termination – Release factor binds to the stop codon
Initiation steps (Prokaryotes)
Small subunit and first tRNA assemble (called the initiation complex)
initiation complex assembles around the start codon with a Shine-Dalgarno sequence before it
Large subunit assembles on top of the first tRNA in the P site
Initiation steps (Eukaryotes)
Small subunit and first tRNA assemble (called the initiation complex)
Initiation complex attaches to the 5’ cap of the mRNA and moves in the 3’ direction until it reaches the first start codon
Large subunit assembles on top of the first tRNA in the P site
Elongation
New tRNA enters the A site
Peptide bond forms between the amino acids in the A and P site
tRNA in the P site releases its amino acid
Ribosome shifts over one codon
tRNA that is now in the E site leaves
Repeat
Termination Process
Stop codons are the only codons that do not have a tRNA that are complementary to them
The are recognized by a protein called release factor
Ribosome shifts and there is now a stop codon in the A site
Release factor (a protein) enters the A site and binds to the stop codon
Ribosome complex completely breaks apart releasing the polypeptide
phosphorylated, chaperone proteins help fold it, etc.
a ton of things can happen before a polypeptide becomes a protein
Review Question
What can happen to a polypeptide after it is translated?
modRNA (Nucleoside-modified messenger RNA)
Synthetic mRNA made with nucleotide analogues
Nucleotides are not A,U,C, or G
But are read as A,U,C, or G by the ribosome
Used to trick the cell into coding for a specific protein
Moderna and Pfizer COVID-19 vaccine
Why use nucleotide analogues over natural nucleotides?
tba; prob make it easier to get into cell
Review Question
The modRNA in the COVID-19 vaccines are put into microscopic lipid nanoparticle, why?
Initiation complex
The complex of the Small ribosome subunit + first tRNA assembling in initiation (translation)
turn genes on/off
BPQ
Why regulate gene expression?
multicellular has cell specialization → altho every cell is genetically the same, diff genes expressed in diff cells
BPQ
How might regulation in be different in multicellular organism than unicellular organism?
turning genes on/off leads to evolution (changes over time)
BPQ
How might gene expression effect heredity and evolution?
Factors that can affect gene expression
Transcriptional (Prokaryotes & Eukaryotes)
Transcription factors (Pro & Eu)
Activators
Inhibitors/Repressors
Histones (Eu & Archae)
DNA Methylation (Pro & Eu)
Post Transcriptional (Eu)
MicroRNA - miRNA
Alternate splicing
Transcription
Promoter – region where RNA polymerase binds
RNA polymerase creates mRNA from the template strand
Regulatory Sequence
a noncoding region of DNA that can influence gene expression.
Promoter (Pro & Eu)
Operator (Pro)
Enhancer (Pro & Eu)
Silencers (Eu)
Regulatory Gene
a coding gene that produces a product that can influence gene expression
Transcription Factor
a protein that can influence gene expression by binding regulatory sequences.
Activator – turns on / promotes transcription
Repressor – turns off / decreases transcription
Prokaryotic Operon
a series of related genes where transcription is initiated by one promoter
Creates 1 mRNA & multiple proteins
often regulated by a repressor that attaches downstream of the promoter in a sequence called an operator (a regulatory sequence)
Repressor physically blocks RNA polymerase
all the genes in an operon are LINKED
1 gene per promoter allows for complex expression in eukaryotes
BPQ
In Eukaryotes 1 promoter always transcribes 1 gene and only 1 gene. What might be some reasons for this? Why do Eukaryotes not use operons?
Lac Operon (Repressor: Inducible)
Inducible Operator
Lactose is an Inducer that triggers the transcription of genes responsible for the breakdown of lactose
Inducible Operator
transcription is “turned off” until an inducer binds to the repressor
Inducer releases the repressor from the operator
Lac Operon (Activator)
Activators bind to enhancers (regulatory sequence) and guide RNA polymerase onto the promoter.
cAMP is a coactivator that is produced in the absence of glucose. cAMP “turns on” the activator CAP and triggers transcription of genes responsible for the breakdown of lactose
off
off
off
on
both must be “on” for Lac Operon to turn on
BPQ
Determine if the Lac Operon is turned “on” or “off”
Glucose present, Lactose present
Glucose absent, Lactose absent
Glucose present, Lactose absent
Glucose absent, Lactose present
Trp Operon (Repressor: Repressible)
Repressible Operator
Tryptophan is a corepressor that stops the transcription of genes responsible for the synthesis of tryptophan
Repressible Operator
transcription is “turned on” until a corepressor binds to the repressor.
Corepressor enables the repressor to bind to the operator
lactose comes from an inducible operator - off by default; lactose presence turns on the lactase gene
tryptophan comes from a repressible operator - on by default; trp presence turns off the gene bc there’s enough trp now
BPQ
Why does Lactose “turn on” gene expression while Tryptophan “turn off” gene expression?
Eukaryote Transcription Factors
Transcription factors
Activators
Inhibitors
Basal/general transcription factors
Regulatory sequences
Enhancer (upstream of the promoter)
Silencer(upstream of the promoter)
Promoter
**unlike the Prokaryotes with their operators, Eukaryotes have no regulatory sequences downstream of the promotor
Basal/General Transcription Factors
All Eukaryotic genes require basal transcription factors for the RNA polymerase to bind to the promoter
RNA Polymerase in prokaryotic cells can often bind on its own
Upstream Regulatory Sequences
Eukaryotic regulator sequences can be very far upstream from the promotor
1 regulator sequence can affect multiple promotors/genes
Prokaryotic regulator sequences are always proximal to the promotor
Can only effect 1 promotor(but multiple gene/see operon)
DNA Methylation
A methyl group can be added to the nitrogen bases C or A (usually C)
Affect promotors
“Turn off” gene expression
Affect Histones
“Turn off or on” gene expression
often leads to more long-term patterns in gene expression
growth hormones for cell specialization
BPQ
Considering the long-term effects of DNA methylation, what is likely a common application of DNA methylation in multicellular organisms?
Post Transcriptions Gene Expression (Eu)
MicroRNAs (miRNA)
Small noncoding RNAs that can bind to mRNA
Prevent translation
Accelerate mRNA breakdown
Alternate splicing
Gene Expression and Cell Differentiation
Multicellular Eukaryotes need to express different genes in different cells
see which genes are turned on/off due to cell specialization (?)
BPQ
A Liver tumor is biopsied and found to be metastatic lung cancer
How can we tell that a tumor in the liver came from the lungs?
Gene Expression and Evolution
Changes in gene expression are often more consequential than changes to the genes themselves
Mutation
change in DNA sequence
good, bad, neutral
mutations in regulatory sequences (e.g. promoters) (affect how MUCH of a protein is made)
BPQ
Can a mutation affect a phenotype w/o affecting coding (transcribed and translated) DNA? how?
Mutation Effects
Nothing - non-coding DNA (junk DNA), introns, 3rd nucleotide in codon
AA seq. - coding DNA
Protein Output - Reg. seq. & reg. genes
Mutations in Coding DNA
Missense Mutation
Silent Mutation
Nonsense
Insertion/deletion
Frameshift
Missense mutation
change 1 nucleotide (any of 3 in codon) to another resulting in AA change
Silent mutation
change 1 nucleotide to another BUT does NOT result in AA change (3rd nucleotide sometimes)
no effect sometimes
Nonsense Mutation
changes AA to stop codon
Insertion/deletion mutation
addition/subtraction of nucleotide
If 3, 6, 9, etc. nucleotides are added, a frameshift mutation does NOT occur
Frameshift Mutation
changes all AA downstream when the insertion/deletion is not divisible by 3
3rd nucleotide b/c most often causes silent mutations that tend to have no effect
BPQ
In a coding gene found across multiple species, where would we most likely find differences in nucleotides?
First nucleotide of a codon
Second nucleotide of a codon
Third nucleotide of a codon
Chromosomes (Human vs Chimpanzee)
Human – 46 Chromosomes (22 autosomes, 2 sex chromosomes)
Chimpanzee – 48 Chromosomes (23 autosomes, 2 sex chromosomes)
in chromosome 2, either humans put the two chimp chromosomes tgt, or chimps split them apart after we diverged
but overall the genes are the same, the chromosomes in 2 are just split (shows we are closely related!)
Karyotype
image of all the chromosomes in a single cell
M phase (?)
Review Question
What stage of the cell cycle does the karyotype show
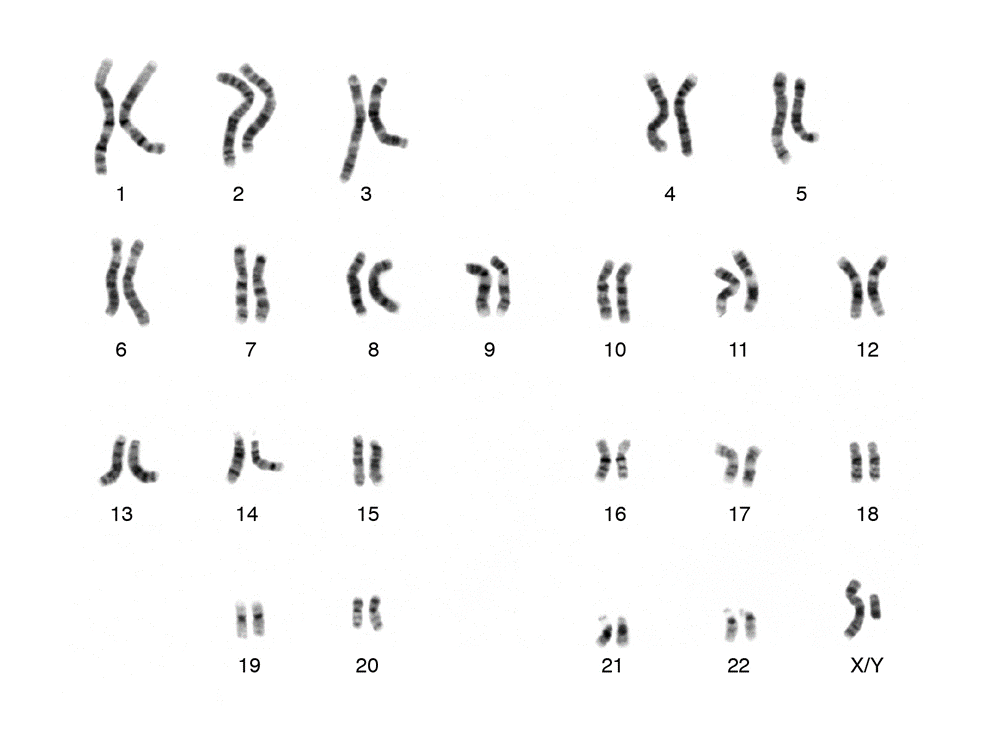
Extra/Missing Chromosomes
Euploid
Aneuploid
Polyploidy
Euploid
having correct number of chromosomes (humans = 46)
Aneuploid
addition or loss of a chromosomes (1 pair)
Monosomy - missing a chromosome from a pair
Trisomy - having an extra chromosome in a pair