Genetics, variation and independence
1/84
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
85 Terms
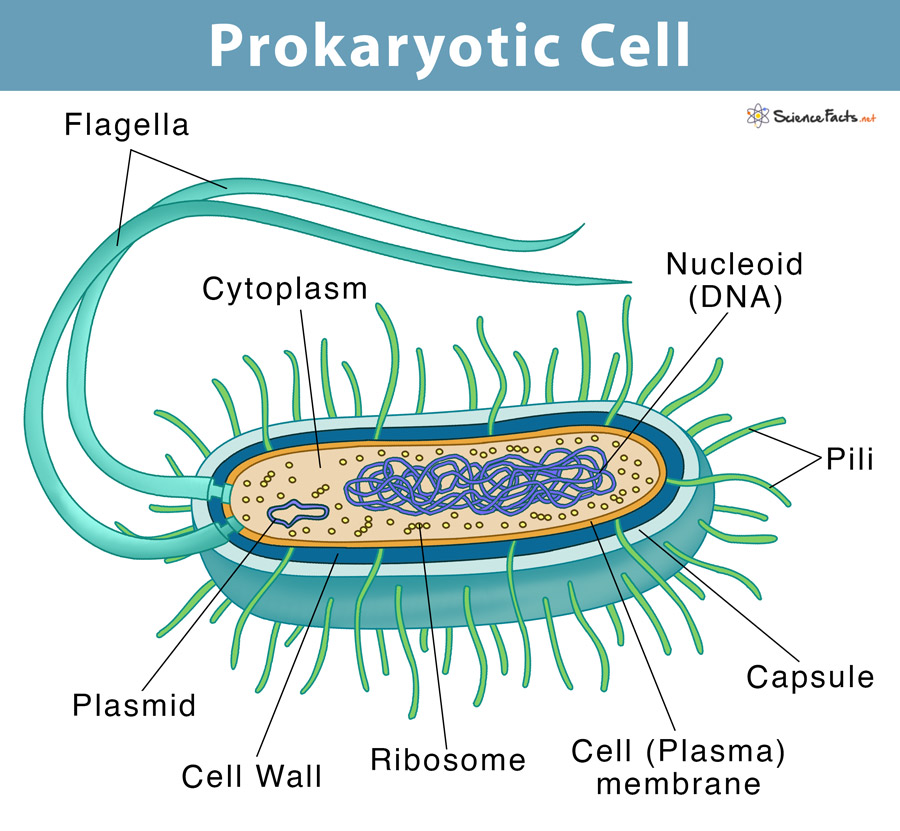
Prokaryotic DNA
single-stranded
single, circular loop of DNA = nucleoid (not a chromosome)
1 or more plasmids
small circular DNA molecules
can be exchanged between cells
usually only contain a few genes
plasmids > accessible for proteins required for gene expression and therefore contain genes required often, quickly/ in emergencies
contain genes for antibiotic resistance
Eukaryotic DNA
Eukaryotic chromosomes
human cells have 23 pairs
haploid (n) = cell/ nucleus containing single copy of each chromosome
diploid (2n) = cell/ nucleus containing 2 sets of chromosomes/ 2 copies of each chromosome
no of chromosomes diff for all organisms
loci = position of gene on chromosome
Gene
= base sequence of DNA that codes for the amino acid of a polypeptide/ a functional RNA (e.g. tRNA/ rRNA)
allele = alternative form of a gene
shape and behaviour of protein molecule depends on exact sequence of these amino acids (i.e. primary structure)
genes in DNA control protein structure
Non-coding DNA
genome contains many non coding sections of DNA (doesn’t code for amino acids)
found between genes, as non-coding multiple repeats
means contain same base sequence repeated multiple times
can be found within genes: introns
coding exons separated by 1 or more introns
during transcription, eukaryotic cells transcribe whole gene (all introns and exons) to produce pre-mRNA molecules
before pre-mRNA exits nucleus, introns removed and exons (coding sections) joined in process = splicing
Proteome
full range of proteins produced by genome
usually large amount of post-translational modification of proteins (often in Golgi)
each gene also capable of producing multiple different proteins via alternative splicing
genome = complete set of genes (e.g. in a cell)
but not every gene expressed in every cell, depends on cell type
DNA
double
long
double helix
deoxyribose
AT CG
found in nucleus
constant (no variability of quantity)
very stable (double stranded)
mRNA
single
short
single strand
ribose
AU CG
found in cytoplasm
variable with level of protein synthesis
least stable
tRNA
single
short
clover-leaf as forms H bonds between complementary bases within strand
ribose
AU CG
found in cytoplasm
variable with level of protein synthesis
more stable than mRNA
anticodon = 3 bases complementary to codon on mRNA
amino acid specific to anticodon
transcription
DNA helicase breaks H bonds to unzip DNA
1 template strand used to produce mRNA molecule by complementary base pairing with complementary base pairing with free RNA nucleotides
RNA polymerase joins nucleotides with phosphodiester bonds
DNA strand rejoins behind
mRNA leaves nucleus through nuclear pore
Post-transcriptional modification
Splicing
only eukaryotes, mRNA produced by transcription is modified inside nucleus, before translation
immediately translation for prokaryotes
Alternative splicing
exons can be spliced in many diff ways to produce diff mature mRNA molecules
so single eukaryotic gene can code for more than 1 polypeptide chain
(so proteome bigger than genome)
translation
mRNA binds to ribosome
ribosome moves to start codon
tRNA brings specific amino acid
anticodon on tRNA binds to complementary codon on mRNA
ribosome moves along to next codon
amino acid joined by peptide bonds by condensation reactions using ATP
continues until stop codon reached, mRNA molecule complete
destination of proteins syntehsised on
free ribosomes (in cytoplasm) for polypeptides for internal cell use
ribosomes bound to RER for use in lysosomes/ secretion
triplet code
sequence of 3 DNA bases that codes for specific amino acid, or is stop/start codon
codon
sequence of 3 mRNA bases that codes for specific amino acid
anticodon
sequence of 3 tRNA bases complementary to codon
no. of diff amino acids
20 amino acids, 64 different triplet bases
stop codons
don’t code for amino acids, causes ribosome and mRNA to detach
start codon
codes for amino acid Met
causes ribosome and mRNA binding
degenerate
more than 1 mRNA codon / triplet codon code for same amino acid
universal
codons code for same amino acids in all species
non-overlapping
each base only read once in codon part of
gene mutations
= change in nucleotide base sequence of DNA
DNA base sequence determines primary structure, can lead to changes in polypeptide that gene codes for
Deletion (+insertion) of nucleotides
changes amino acid coded for
knock-on effect by changing groups of 3 bases further on in DNA sequence
frameshift mutation
may dramatically change amino acid sequence produced from this gene and ability of polypeptide to function
substitution of nucleotides
only change amino acid for triplet where mutation occured
silent mutations
doesn’t alter amino acid sequence of polypeptide and protein function
degenerate nature
missense mutations
alters single amino acid
change in tertiary structure: change in type and position of bonds between R groups
nonsense mutations
creates premature stop codon
signal for cell to stop translation of mRNA molecule into amino acid sequence
short and incomplete polypeptide chain affecting final protein structure
causes of mutations
can arise spontaneously during DNA replication
rare but occur at predictable frequency
mutagenic agents
high ionising radiation can disrupt DNA structure
chemicals e.g. NO2 can directly alter DNA structure/ interfere with transcription
mutation passed onto offspring if
mutation occurs in cell that will ‘divide into’ a gamete e.g. ovary/ testes
chromosome mutations
non-disjunction occurs when chromosomes fail to separate during meiosis
occurs spontaneously
gamete may end up with extra pair or no copies of particular chromosome
= diff no. of chromosomes
if abnormal gametes take part in fertilisation, chromosome mutation occurs as diploid cell will have incorrect no. of chromosomes
2 types of chromosome mutation
change in whole set of chromosomes (structure)
change in no. of individual chromosomes
meiosis
produces genetically diff daughter cells
meiosis I
homologous chromosomes pair up and separate
meiosis II
chromatids separate and gametes formed, each with half original no. of chromosomes
independent assortment
how homologous chromosomes pair up on spindle in metaphase I
random
alleles of 2/more diff genes get sorted into gametes
how homologous chromosomes line up in random orientations at middle of cell
independent segregation
how homologous pairs separate in anaphase I
2²³ possible combinations of chromosomes per human gamete
independent crossing over
metaphase I
homologous pairs = bivalent
chromosomes break, exchange DNA between non-sister chromatids and rejoin
= recombinant chromosomes = new combination of alleles
importance of meiosis
produce haploid gametes
important to maintain diploid number at fertilisation
results in genetically varied gametes, with novel allele combinations
how meiosis results in genetic variation
independent segregation of homologous chromosomes
crossing over
random fusion of gametes during fertilisation
= unique combination of alleles
mitosis
mitosis
produces 2 daughter cells per cycle (diploid)
produces genetically identical daughter cells
unless mutation occurs during DNA replication
1 nuclear division per cycle
meiosis
produces 4 haploid cells, genetically varied through
crossing over to produce new combinations of alleles
independent segregation of homologous chromosomes
2 nuclear divisions per cycle
take place in testes, ovaries, anthers
genetic diversity
no. of different alleles of genes in a population
genetic variation transferred down generations and results in genetic diversity
new allele may be dis/advnatageous or have no effect on phenotype
new alleles may remain hidden within population for several generations before contribute to phenotypic variation
gene pool
all of genes and their diff alleles present in an interbreeding population
effect of genetic diversity
genetic diversity → allows NS → adaption
diff alleles → diff phenotypes within population
environmental factors act as selection pressure
biotic/ abiotic
increase chance of individuals with specific, favoured phenotype to survive and reproduce (have higher fitness)
small gene pool = < likely to be able to adapt to changes in environment and so become vulnerable to extinction
higher fitness
= able to survive and pass allels to offspring
Discontinuous variation
genetic factors
characteristics controlled by single gene, not influenced by environmental factors
few distinct forms, no intermediate types
e.g. ABO blood groupings
Continuous variation and environmental influences
e.g. environmental factors limiting height of buttercup plant
biotic (competition, pests, pathogens)
abiotic (light intensity, temp, wind speed, humidity)
continuous variation due to polygenes
many characteristics controlled by multiple genes and environment
leads to normal distribution curve on graph
principle of natural selection
random mutation can result in new alleles of gene
individuals with advantageous allele more likely to survive, reproduce and pass on their allele to next generation
over many generations, new allele increase in frequency in population
(sexual selection, those with advantegeous allele more likely to reproduce, NOT survival)
selection
= process by which organisms that are better adapted to their environment are > likely to survive and reproduce
selection pressure
= environmental factors that affect chance of survival of organism
stabilising selection
keeps allele frequency constant over generations
preserves average phenotype
selects for 1 phenotype and selects against extreme phenotypes and so against evolutionary change
occurs when environment constant
variation decreases, mean constant
e.g. birth weight, no. of offspring, cactus spine density
directional selection
occurs in changing environments
when there is clear advantage in population changing in 1 particular direction
combination of alleles at 1 extreme of phenotype selected against and those at other end selected for
variation constant, mean shifts in 1 direction
e.g. development of antibiotic resistance in bacteria, peppered and melanic forms of peppered moth
disruptional selection
results in polymorphism
can lead to speciation
extremes selected for
mean selected against
e.g. long fur and active in winter, short fur and active in summer
anatomical adaptations
structural / physical feature
e.g. white fur of polar bears provides camouflage in snow so has < chance of being detected by prey
physiological adaptations
biological processes within organism
e.g. mosquitos produce chemicals that stops animal’s blood clotting when they bite, so can feed more easily
behavioural adaptations
way organism behaves
e.g. cold-blooded reptiles bask in sun to absorb heat
effect of adaptation and selection on population
evolution = change in adaptive features of population over time as result of natural selection
over generations, those features better adapted to environment become > common
= whole population > suited
if 2 populations of species isolated from each other and become so diff in phenotype that can no longer interbreed. to produce fertile offspring = 2 new species
→ accumulated genetic differences
evolution drives speciation
species
group of organisms with similar characteristics which can interbreed to produce fertile offspring
issues with species definition
asexual organisms don’t interbreed
extinct species, only have fossil records
some species interbreed to produce fertile offspring (often not well adapted)
ring species: species with geographic barrier and form ring
still have gene flow but can only interbreed with groups closest to them
courtship
enables organisms to
female selection of advantageous alleles (sexual selection) and resources
recognise members of own species
identify mate that is capable of breeding
form pair bond
synchronise mating
for external fertilisation, to release sperm and egg at same time to maximise chance of them meeting
become able to breed
importance of classification
common name for species
group organisms with shared characteristics
how closely related organisms are
binomial system of classification
genus (shared with all closely related species)
species (unique type of organism)
phylogenetic classification
attempts to arrange species into groups based on evolutionary origins and relationships
uses hierarchy in which smaller groups placed within larger groups, no overlap
Kingdom
Phylum
Class
Order
Family
Genus
Species
keep
ponds
clean
or
frogs
get
sick
archaea
most live in extreme environments
no nucleus so prokaryotic
unicellular (single-celled)
70s ribosomes
proteins synthesis more similar to eukaryotes, more complex form of RNA polymerase than bacteria
circular ‘chromosomes’
histones
cell wall not murein/peptidoglycin
fatty acid chains attached to glycerol by ether linkages
bacteria
no membrane-bound organelles
unicellular
70s
DNA: circular, shorter, no chromosomes
no histones
cell wall made of murein
fatty acid chains atached to glycerol by ester bonds
eukarya
membrane-bound organelles e.g. nucleus and mitochondria
multi/uni
80s
DNA: double helix, linear, arranged into chromosomes
histones
cell wall: plants - cellulose, fungi - chitin
fatty acid chains attached to glycerol by ester bonds
artificial classification
divides organisms according to morphological features
can be analogous: same function but not evolutionarily related e.g. wings
phylogenetic classification
based on evolutionary relationships between organisms and shared features derived from common evolutionary ancestry
homologous characteristics
phylogeny = evolutionary relationships between organisms
genome sequencing for evolutionary relationships
using DNA/ mRNA/ amino acids of a protein
scientists choose specific proteins/ sections of genome for comparison between organisms
looking at multiple proteins/ regions of genome = > accurate
NB protein needs to present in wide range of organisms and show sufficient variation between species
> similar sequence = > closely related
= separated species > recently
separated for longer = > time to accumulate mutations and changes to their DNA, mRNA and amino acid sequences
immunology
to compare proteins of organisms
albumin = protein found in many species
Methodology
pure albumin samples extracted from blood samples taken from multiple species
each sample injected into diff rabbit
each rabbit produces antibodies fro that specific type of albumin
diff antibodies extracted from diff rabbits and are mixed with diff albumin samples
ppt (antibody-antigen complexes) resulting from each mixed sample is weighed
Results
> weight of ppt = > degree of complementarity between antibody and albumin
e.g. antibodies produced against human albumin will produce > amount of ppt when exposed to chimpanzee albumin than rat albumin as > closely related
species richness
no. of diff species in community
community = group of populations of diff organisms living in same place at same time that interact with each other
can be misleading as doesn’t take into account abudnance
species evenness
no. of individuals in each species (abundance)
biodiversity important because
allows adaptation to changing conditions e.g. climate change, new pathogen)
food webs and symbiotic relations (e.g. pollinators)
medicines
ecosystem/ habitat diversity
range of habitats in particular area/ region
> diff habitats = > biodiversity
e.g. coral reef with lots of microhabitats and niches to be exploited
1/ 2 diff = < biodiversity
e.g. large sandy deserts as same conditions in whole area
species diversity
richness and evenness
> = > stable as > resilient to environmental change
doesn’t have dominant species
index of diversity
looks at evenness and richness
describes relationship between no. of species present and how each species contributes to total no. of organisms present in that community
d = N(n-1)/ ∑n(n-1)
n = total no. of organisms for single species in community
N = total no. of organisms in community (all species)
> num = > biodiversity
impact of farming on biodiversity
fertilisers: eutrophication
selective breeding
pesticides: kill pests
kill pollinators and other species
herbicides kill weeds and other plants
hedgerow removal
pollinators and birds and others live on hedgerows
deforestation
filling in ponds, draining marshland (habitat)
monocultures: 1 type of plant grown on fields
supports fewer species
decrease biodiveristy
conservation methods in farming
maintain ponds
crop rotation
grow nitrogen-fixing crops e.g. legumes to replace nitrates
limit/ regulate use of
pesticides → use biological controls e.g. natural predators)
fertilisers → use organic = manure (still causes eutrophication)
herbicides
conserve/ replant
hedgerows
field margins e.g. wild flowers
natural meadows
don’t cut grass until after flowering
biodiversity vs profit
high yield + profit = 2 factors that make farming viable
difficult to find balance between conservation and farming
practices to increase biodiversity = expensive, labour and time intensive
genetic isolation
2 groups reproductively isolated → genetically isolted
changes that occur in allele freq of each group not shared so evolve independently which can lead to 2 diff species
can’t successfully interbreed
investigating genetic diversity
now:
displays of measurable characteristics
nucleotide base seq of DNA/ proteins
originally
comparison of observable characteristics to suggest evolutionary relationships (could be analogous)
investigating diversity using mtDNA
zygote only contains mitochondria of egg so only maternal mtDNA present in zygote
= no crossing over so base seq can only be changed by mutation
so scientists can research origin of species, genetic drift and migration events
investigating diversity using mRNA analysis and comparison
diversity in mRNA bases will be complementary to base seq of DNA after transcription
mRNA = easier to isolate as found in cytoplasm and there are usually multiple copies of same mRNA
can still give comparison between diff species
mRNA = only coding regions
investigating diversity using amino acid seq analysis and comparison
protein easier to isolate than DNA
amino acid seq of proteins evolve much slower than DNA, especially if protein of high importance
likely closely related species will have same amino acid seq even though split from common ancestor millions of yrs ago
bc shape and function and specificity of protein determined by amino acid sequence as position of amin o acid determines IMF between R groups
phylogenetic tree
conflicting evidence for relationships between organisms e.g. primates(order) illustrates need to use variety of evidence from diff sources in drawing valid scientific conclusions