3.1 - BIOLOGICAL MOLECULES
1/140
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
141 Terms
Polymers
Large, complex molecules made of long chains of monomers

Monomers
Small, basic molecular units
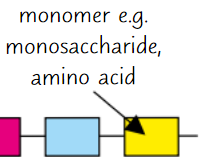
Monomer examples
Monosaccharides, amino acids, nucleotides
Carbohydates contain ____
C, H, O
Monosaccharide examples
Glucose, fructose, galactose
Carbohydrates are (mono/polysaccharides)
Polysaccharides
Glucose
Hexose sugar - 6 C atoms per molecule
Has 2 isomers - alpha (α) and beta (β)
α-glucose structure
(alpha - H is on top)
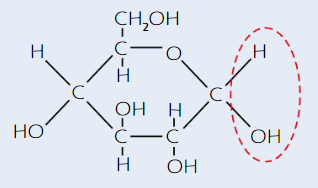
β-glucose structure
(beta - H is on bottom)
Condensation reaction
Two molecules join with formation of new chemical bond
+ water molecule released
Monosaccharides are joined by ____
condensation reactions
Glycosidic bond
Bond between two monosaccharides
Sucrose
Disaccharide formed by glucose + fructose (via condensation reaction)
Lactose
Disaccharide formed by glucose + galactose (via condensation reaction)
Maltose
Disaccharide formed by glucose + glucose (via condensation reaction)
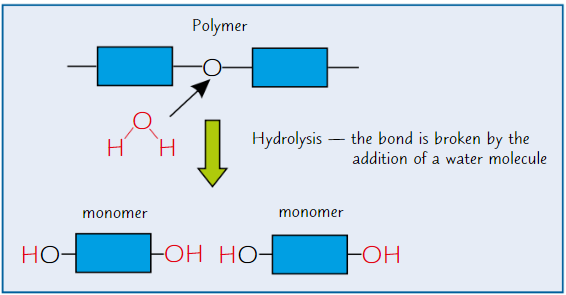
Hyrolysis reactions
Breaking of chemical bond between monomers using water molecule
What is starch made of?
Mixture of amylose + amylopectin - polysaccharides of α-glucose
Plants store excess glucose as ___
starch
Why is starch good for storage?
Insoluble in water → doesn’t affect water potential
→ doesn’t cause water to enter cells by osmosis
→ good for storage
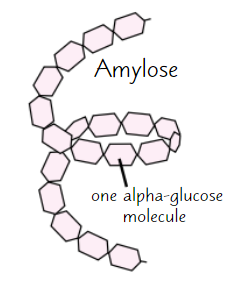
Amylose
Long, unbranched chain of α-glucose
Angles of glycosidic bonds → coiled structure
→ compact
→ good for storage
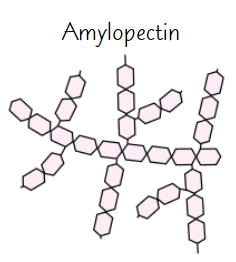
Amylopectin
Long, branched chain of α-glucose
Side branches allow enzymes that break down molecule to reach glycosidic bonds easily
→ glucose can be released quickly
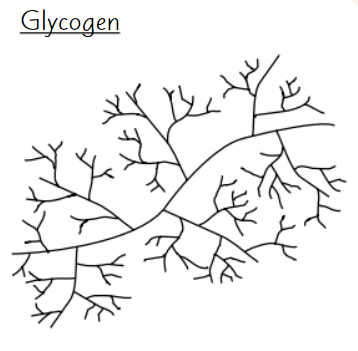
Glycogen
Polysaccharide of α-glucose
Animals store excess glucose as glycogen
Glycogen structure
Highly branched structure
→ stored glucose can be released quickly
Compact
→ good for storage
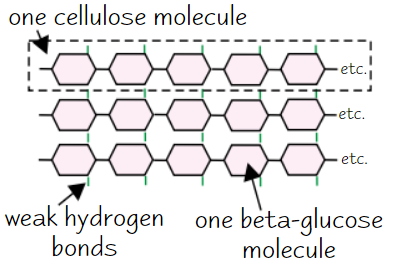
Cellulose
Made of long, unbranched chains of β-glucose
When β-glucose molecules bond, they form straight cellulose chains
Cellulose chains linked by H bonds to form microfibrils (strong fibres)
→ strong fibres make cellulose a good structural support for cells
Which sugars are reducing sugars?
All monosaccharides + some disaccharides (e.g. maltose, lactose)
Test for reducing sugars
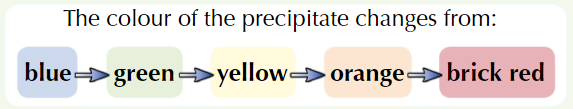
Add Benedict’s reagent (blue) to sample + heat in water bath at 100ᵒC
If positive test, coloured ppt forms
Higher conc. of reducing sugar = further colour change
Test for non-reducing sugars
If reducing sugar test negative, must do non-reducing sugar test
Get new sample of test solution, add dilute hydrochloric acid + heat in water bath at 100ᵒC
Add sodium hydrogencarbonate to neutralise
Carry out Benedict’s test (reducing sugar test)
If positive test, coloured ppt forms
If negative, solution stays blue → no sugar in solution
Test for starch
Add iodine dissolved in potassium iodide solution to sample
If positive test, sample changes from browny-orange → blue-black
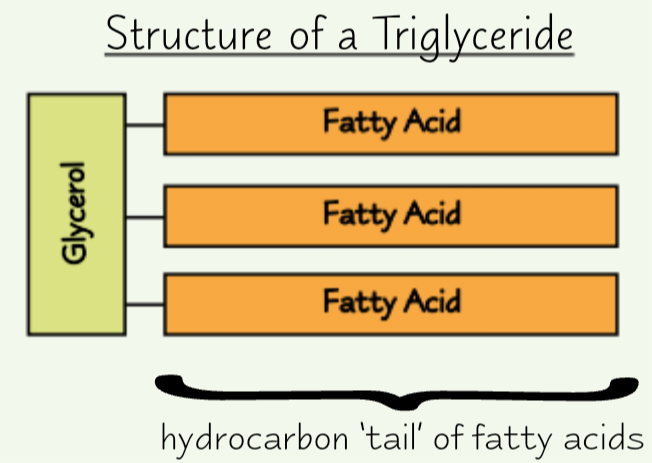
Triglyceride structure
1 glycerol + 3 fatty acid
Fatty acid molecules
Have ‘tails’ made of hydrocarbons
Tails ‘hydrophobic’ (repel water)
→ lipids insoluble in water
All fatty acids have same basic structure, but hydrocarbon tail varies
Basic structure of fatty acid
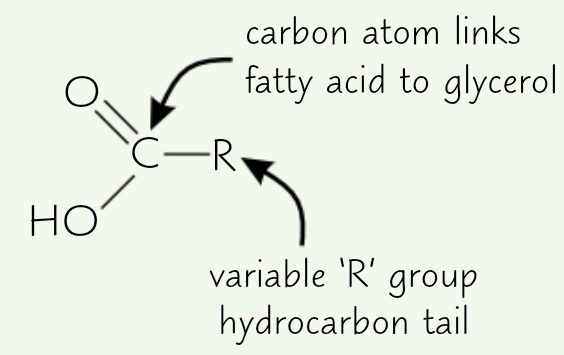
What bond forms between glycerol + fatty acid?
Ester bond
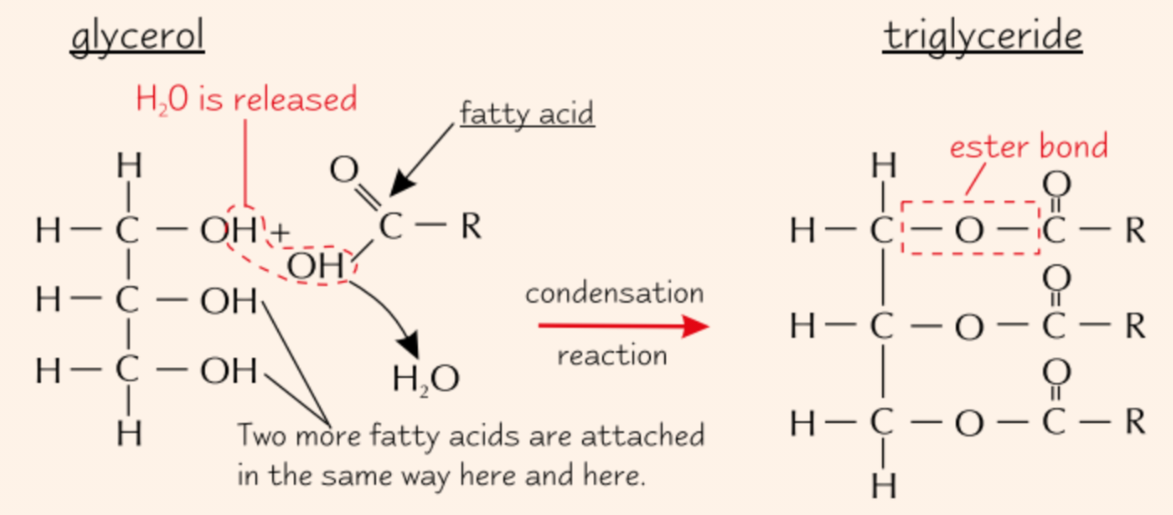
Formation of triglycerides
Ester bond forms between each fatty acid and the gycerol molecule
Each time, water molecule is released
3x condensation reactions
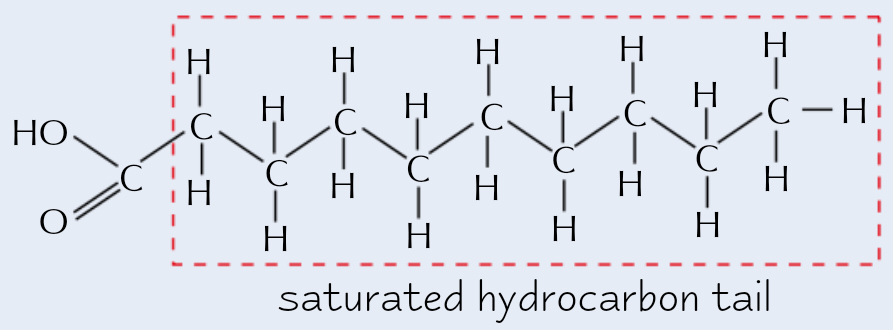
Saturated fatty acid
No double bonds between C atoms (in hydrocarbon tail/R group)
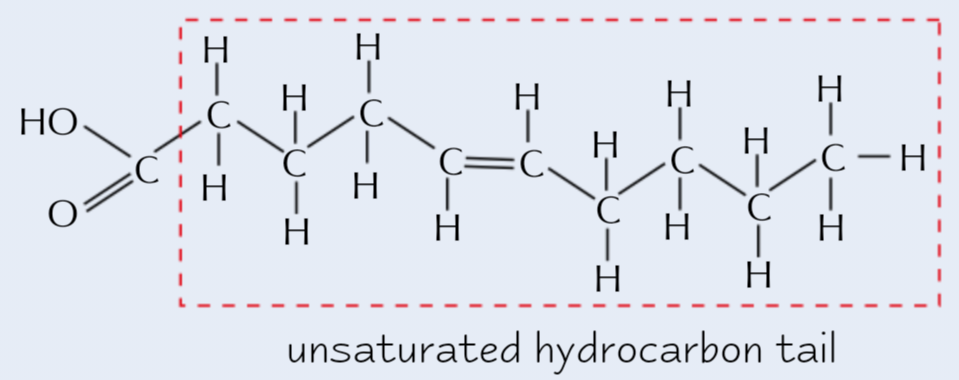
Unsaturated fatty acid
1+ double bond between C atoms
→ causes kinks in chain
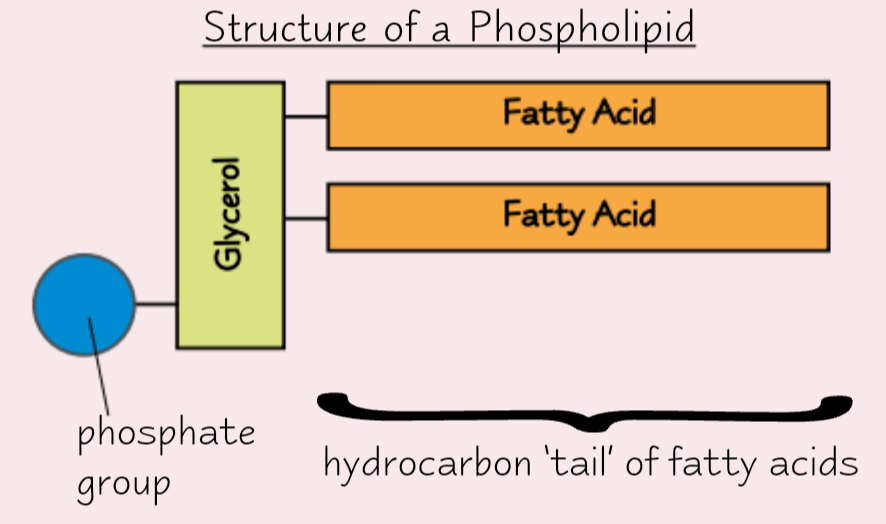
Phospholipids
Found in cell membrane
1 fatty acid replaced by phosphate group
Phosphate group hydrophilic (attracts water)
Structure + function of triglycerides
Energy storage molecules
Long hydrocarbon tails contains lots of chemical energy
→ lots of energy released when they’re broken down
Lipids have 2x energy per gram than carbohydrates
Insoluble
→ don’t affect water potential and cause water to enter by osmosis
Triglycerides stay together as insoluble droplets in cells
→ fatty acid tails hydrophobic so face inwards, shielded from water
Structure + function of phospholipids
Make up bilayer of cell membranes (which control what enters/leaves cell)
Hydrophilic heads + hydrophobic tails
→ form double layer with heads facing out towards water
Centre of bilayer hydrophobic
→ water-soluble substances can’t pass through easily
→ membrane is barrier to those substances
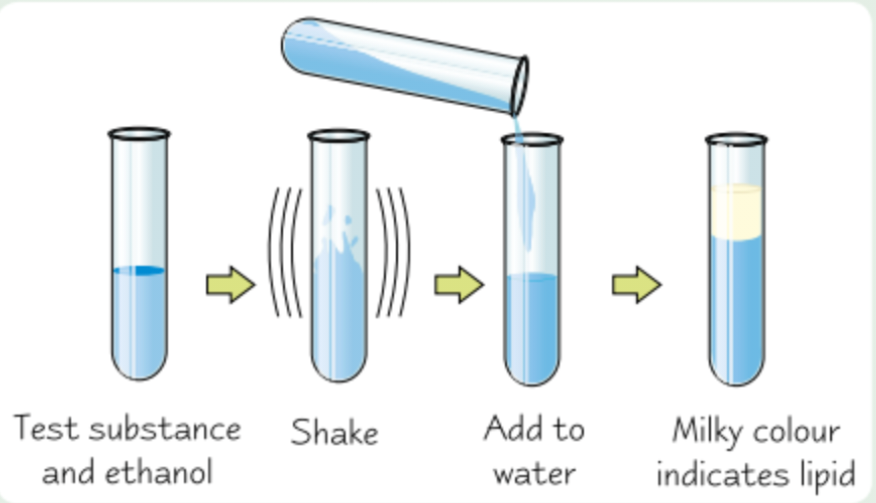
Name of test for lipids
Emulsion test
Test for lipids
Shake test substance with ethanol for 1min so it dissolves, then pour solution into water
Any lipid shows up as milky emulsion
More lipid = more noticeable milky colour
What are the monomers of proteins?
Amino acids
Dipeptide
2 amino acids joined together
Polypeptide
3+ amino acids joined together
Proteins are made of…
One or more polypeptides
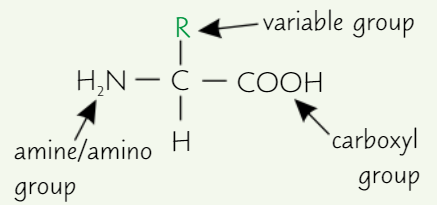
Structure of amino acid
Same general structure:
Carboxyl group (-COOH)
Amino group (-NH₂)
R group (variable group)
How many amino acids are there?
20 - all living things share a bank of 20 amino acids
Differences in amino acids
Only difference is R group
Formation of polypeptides
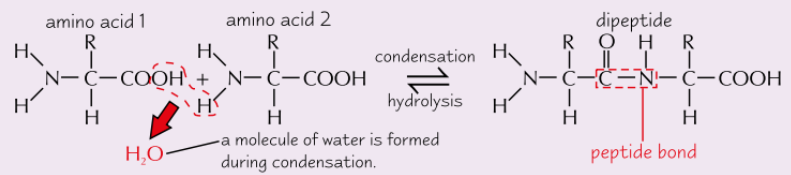
What is the name of the bond between amino acids?
Peptide bond
What is released during condensation reaction?
Water molecule
Primary structure of proteins
Sequence of amino acids in polypeptide chain
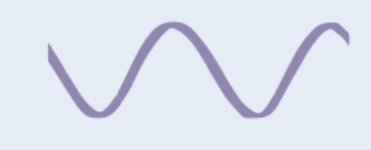
Secondary structure of proteins
Polypeptide chain isn’t flat + straight
H bonds form between amino acids in chain
→ chain coils/folds automatically
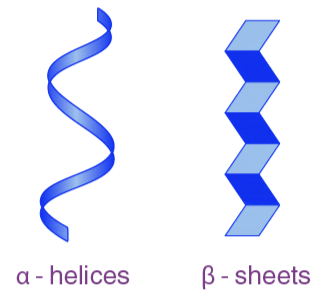
Types of secondary structure in proteins
Can coil into alpha (α) helix
or fold into beta (β) pleated sheet
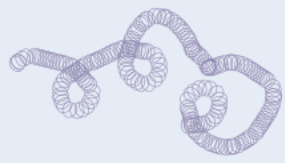
Tertiary structure of proteins
Coiled/folded amino acid chan is coiled/folded further
More bonds form between different parts of polypeptide chain, incl. H bonds + ionic bonds (attractions between -ve and +ve charges on different parts of molecule)
For proteins made from single polypeptide chain, tertiary structure forms their final 3D structure
Example of tertiary structure in proteins
Disulfide bridges form whenever 2 molecules of amino acid cysteine come close together - S atom of one cysteine bonds to S atom of other
Quaternary structure of proteins
Some proteins made of several different polypeptide chains held together by bonds
Quaternary structure is the way polypeptide chains are assembled together
For proteins made from 1+ polypeptide chain (e.g. haemoglobin, insulin), quaternary structure = proteins final 3D structure
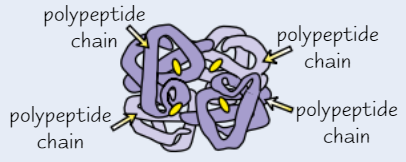
Function of proteins as enzymes
Usually a roughly spherical shape due to tight folding of polypeptide chains
Soluble - often have roles in metabolism, e.g. some enzymes break down larger food molecules
Some enzymes help to synthesise large molecules
Function of proteins as antibodies
Involved in immune response
Made of 2 light (short) polypeptide chains + 2 heavy (long) polypeptide chains bonded together
Have variable regions - amino acid sequences in these regions vary greatly
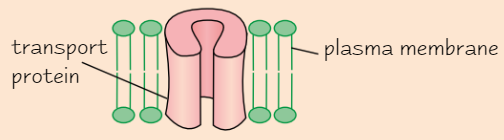
Function of proteins as transport proteins
e.g. channel proteins in cell membranes
Channel proteins contain hydrophobic + hydrophilic amino acids
→ protein folds up to form channel
Transport molecules + ions across membranes
Function of proteins as structural proteins
Physically strong
Have long polypeptide chains lying parallel to each other with cross-links between them
Include keratin (in hair + nails) and collagen (in connective tissue)
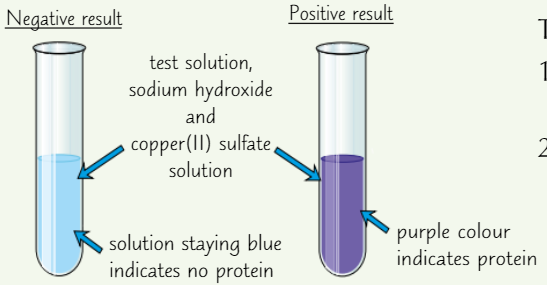
Test for proteins
Add drops of sodium hydroxide solution (solution must be alkaline)
Add copper(II) sulfate solution
Protein present = purple solution
No protein = solution stays blue
Enzymes are known as…
biological catalysts
Enzymes are what type of biological molecule?
Proteins
Enzymes
Catalyse metabolic reactions
at cellular level (e.g. resp.) and for organism as a whole (e.g. digestion)
Can affect structures (e.g. involved in production of collagen) and functions (e.g. resp.)
Intracellular (within cells) / extracellular (outside cells)
Why are enzymes specific?
Have active site with specific shape
Active site = part of enzyme where substrate molecules bind to
Highly specific due to tertiary structure
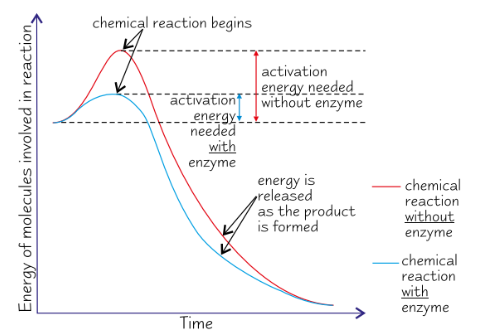
Activation energy
Energy that must be supplied for reaction to start - often provided as heat
How do enzymes catalyse reactions?
Lowering activation energy
→ reactions occur at lower temp.
→ increases RoR
Why does formation of enzyme-substrate complex lower activation energy?
If two substrate molecules need to be joined:
→ being attached to enzyme holds them close together
→ reduces repulsion between molecules
→ can bond more easily
If enzyme is catalysing breakdown reaction:
→ fitting into active site puts strain on bonds in substrate
→ substrate molecule breaks up more easily
Induced fit model
Substrate doesn’t only have to be right shape to fit active site, also has to make active site change shape in the right way
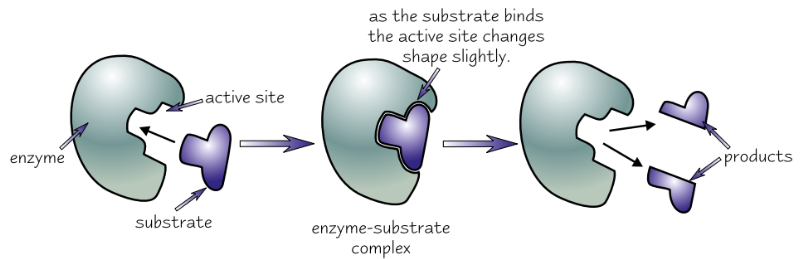
Enzyme’s tertiary structure
Enzymes very specific - usually only catalyse one reaction
because only one complementary substrate fits into active site
Active site shape is determined by enzyme’s tertiary structure (which is determined by primary structure)
Each enzyme has different tertiary structure → different shaped active site
If tertiary structure is altered in any way, shape of active site changes
→ substrate won’t fit in active site → no enzyme-substrate complex → reaction not catalysed
Tertiary structure may be altered by changes in pH + temp
Primary structure of a protein is determined by ____
a gene
Mutation in gene could change tertiary structure of enzyme
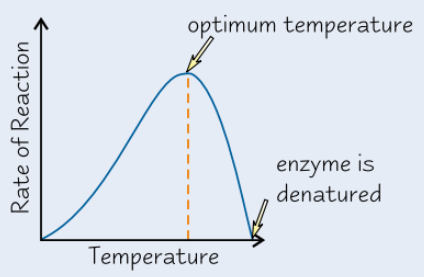
Effect of temperature on enzyme activity
Higher temp → enzyme molecules vibrate more
Temp above certain level → vibrations break bonds that hold enzyme in shape
→ active site changes shape
→ enzyme denatured
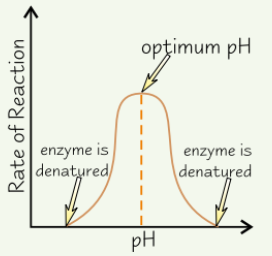
Effect of pH on enzyme activity
Enzymes have optimum pH
Above and below optimum, H⁺ and OH⁻ ions in acids and alkalis damage ionic bonds + H bonds holding enzyme’s tertiary structure in place
→ active site changes shape
→ enzyme denatured
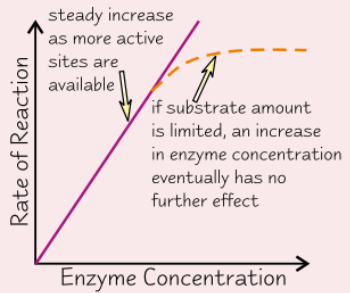
Effect of enzyme concentration on enzyme activity
More enzyme molecules → more likely for substrate molecule to collide and form enzyme-substrate complex
→ increase enzyme conc. = increase RoR
If amount of substrate limited, there is a point where enzyme molecules > substrate → more enzyme = no effect
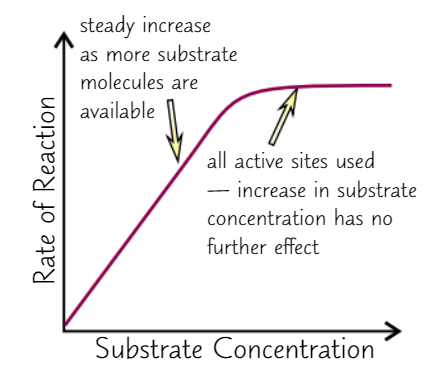
Effect of substrate concentration on enzyme activity
Higher substrate conc. = faster reaction
→ more likely to have collisions
only true up to ‘saturation’ point
all active sites are full → more substrate = no effect
Substrate conc. decreases with time
→ if no other variables change, RoR decreases over time
→ initial RoR is highest
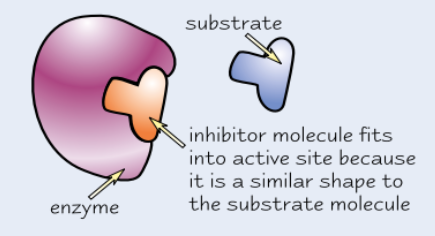
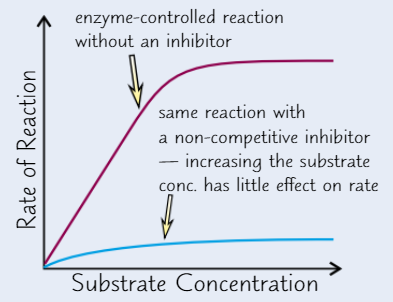
Competitive enzyme inhibitors
Similar shape to substrate molecules
Compete with substrate to bind to active site, but no reaction occurs
Block active site so no substrate molecules can fit
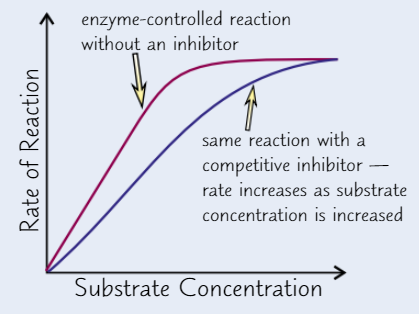
Effect of competitive inhibitor concentration on enzyme activity
High conc of inhibitor → take up nearly all active sites → substrate can’t get to enzyme
Higher substrate conc. → substrate’s chances of getting to active site before inhibitor increase
→ increasing substrate conc → increase RoR (up to a point)
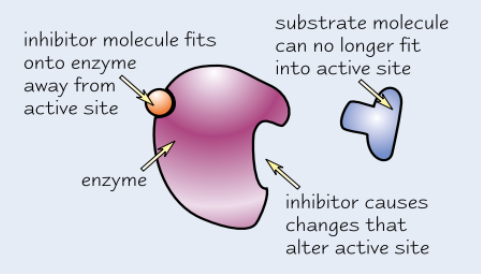
Non-competitive enzyme inhibitors
Bind to enzyme away from active site
→ active site changes shape
→ substrate can’t bind
Effect of non-competitive inhibitor concentration on enzyme activity
Increasing substrate conc. has no effect on RoR - enzyme activity still inhibited
DNA
(deoxyribonucleic acid)
Stores genetic info - instructions needed for organism to grow + develop
RNA
(ribonucleic acid)
One main function is to transfer genetic info from DNA → ribosomes
Ribosomes function
Carry out protein synthesis
Read DNA to make polypeptides (proteins) during translation
What are ribosomes made from?
RNA + proteins
What are the monomers of DNA and RNA?
Nucleotides
Nucleotide structure
A pentose sugar (sugar with 5 C atoms)
A nitrogen-containing organic base
A phosphate group
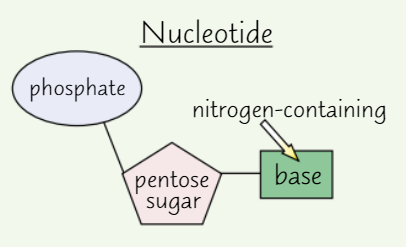
DNA nucleotides
Pentose sugar in DNA nucleotide = deoxyribose
Each DNA nucleotide has same sugar + phosphate group
Base varies
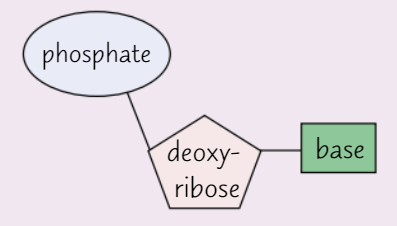
Possible bases in DNA nucleotides
Adenine (A)
Thymine (T)
Cytosine (C)
Guanine (G)
RNA nucleotides
Pentose sugar in RNA nucleotide = ribose
Like DNA, RNA nucleotide has phosphate group + one of four bases
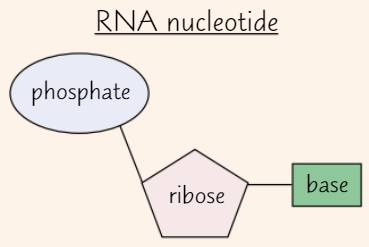
Possible bases in RNA nucleotides
Adenine (A)
Uracil (U)
Cytosine (C)
Guanine (G)
Polynucleotide strand
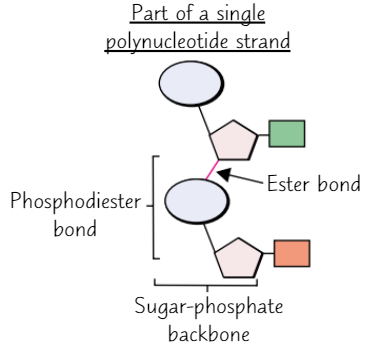
How do nucleotides bond together?
Condensation reaction between phosphate group of one nucleotide + sugar of another
What is the name of the bond between nucleotides?
Phosphodiester bond (consists of phosphate group + 2 ester bonds)
Sugar-phosphate backbone
Chain of sugars + phosphates in polynucleotide
DNA structure
Two DNA polynucleotide strands join by H bonding between bases
Bases join with complementary base pairing
→ always equal amounts A + T and equal amounts C + G in DNA
Two antiparallel (opposite directions) polynucleotide strands twist to form DNA double-helix
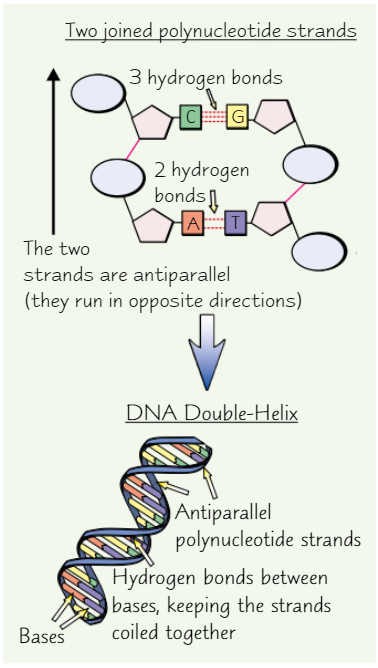
Complementary base pairs in DNA
Adenine with thymine (A-T)
Cytosine with guanine (C-G)
How many bonds form between each base pair?
A-T = 2 H bonds
C-G = 3 H bonds
RNA structure
Single polynucleotide chain
Shorter than most DNA polynucleotides
What was scientist’s previous understanding of DNA?
DNA first observed in 1800s
Scientists at the time doubted it could carry genetic code due to its relatively simple chemical composition
→ thought genetic info was carried by proteins (more chemically varied)
By 1953, experiments showed DNA was carrier of genetic info
Double-helix structure was also discovered that year, by Watson and Crick
Semi-conservative replication
DNA copies itself before cell division → each new cell has full DNA
Half of strands of new DNA molecule are from original DNA molecule
→ genetic continuity between generations of cells
Genetic continuity
Cells produced by cell division inherit their genes from their parent cells