Enzymes + Lysozyme
1/84
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
85 Terms
Why are enzymes important?
they speed up chemical reactions that would otherwise occur too slowly to support life
True or false: Enzymes are highly specific for a particular substrate and also highly specific for the reaction that it catalyzes.
true
refers to the protein component of an enzyme
apoenzyme
refers to the non-protein components of enzymes, usually made up of small inorganic ions (Fe, Mg, Mn) covalently bound to the protein
cofactors
What three inorganic ions are typically cofactors for enzymes?
Fe, Mg, Mn
organic molecules that are often derived from vitamins that act as participants in the catalyzed reaction but are regenerated
coenzymes
Are cofactors organic or inorganic?
inorganic
What are coenzymes usually derived from?
vitamins
Are coenzymes organic or inorganic?
organic
apoenzyme + all cofactors and coenzymes needed for it to become biologically active
holoenzyme
two examples of coenzymes derived from niacin, where the nicotinamide can accept an electron and its 2 associated hydrogens
NAD+, NADP+
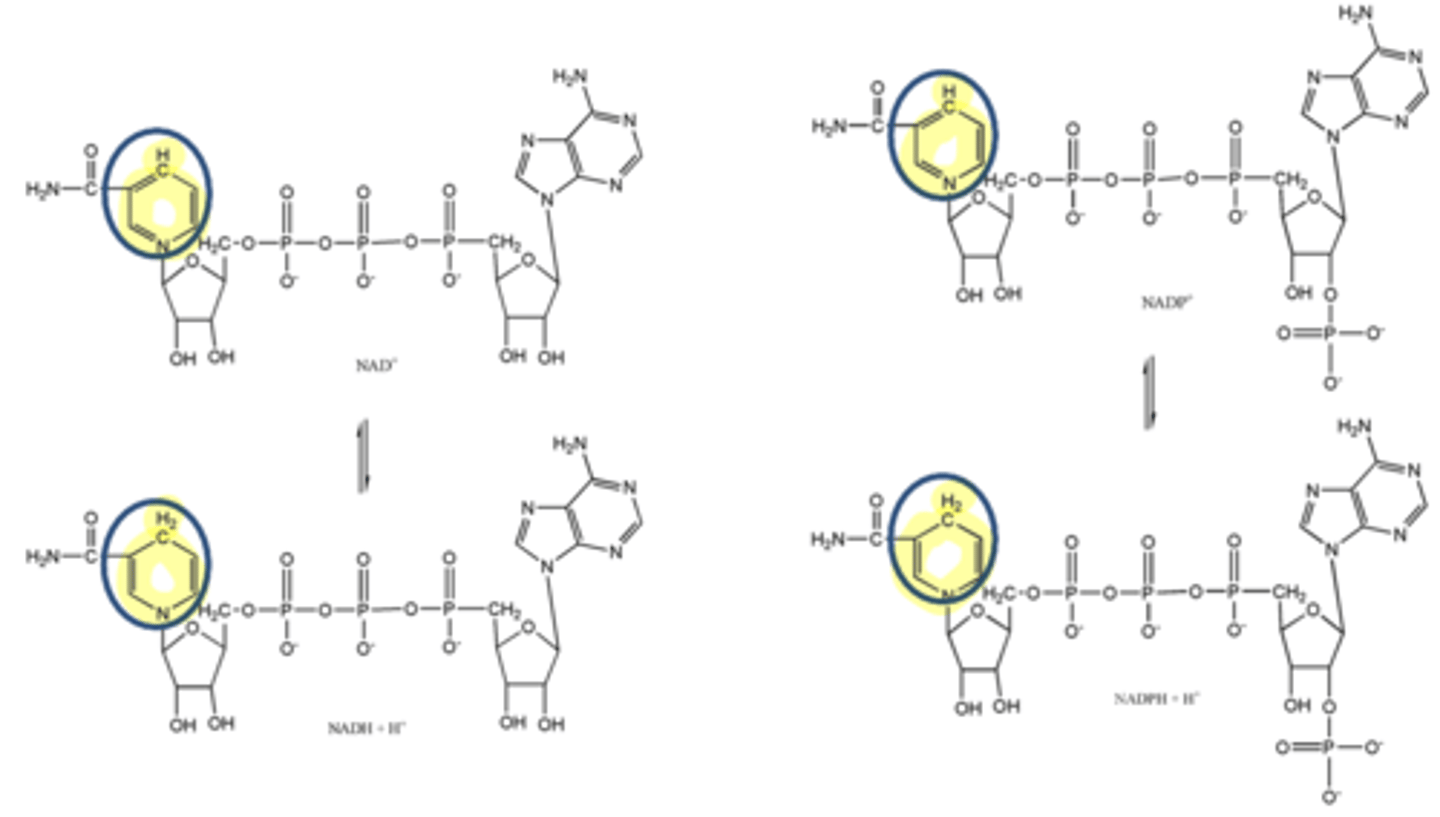
Nicotinamide adenine dinucleotide (NAD+) and nicotinamide adenine dinucleotide phosphate (NADP+) are 2 coenzymes derived from what substance?
niacin
Nicotinamide adenine dinucleotide (NAD+) and nicotinamide adenine dinucleotide phosphate (NADP+) are 2 coenzymes derived from niacin, where the nicotinamide can accept what three things?
electron, 2 hydrogens
enzymes that carry out oxidation-reduction reactions
require cofactors like NAD+ and NADP+
include dehydrogenases
oxidoreductases
example of an oxidoreductase that uses coenzymes (like NAD+ or NADP+) to add or remove hydride ions from a carbon backbone
dehydrogenases
enzymes that transfer functional groups between donor and acceptor molecules
includes kinases
transferases
example of a transferase that phosphorylates substances by transferring a phosphate group
kinase
enzymes that add water across a bond, cleaving the bond
hydrolases
enzymes that break bonds not via hydrolysis or oxidation-reduction
lyases
enzymes that links molecules together
pretty much the reverse of lyases, but technically a type of lyase
synthase
Carbonic anhydrase is an example of what kind of enzyme?
dehydratase
(which is a lyase)
enzymes that carry out conversions between isomers
isomerases
enzymes that catalyze reactions in which two chemical groups are joined with the use of energy from ATP
ligases
place on the enzyme at which the substrate attaches; contains any necessary coenzymes or cofactors
once the substrate attaches, you have an enzyme substrate complex
active site
Is the initial bond between the substrate and enzyme weak or strong?
weak
The binding of a substrate to its enzyme induces a conformational change in the enzyme, optimizing the shape of the active site to allow what to happen? What is created as a result?
allows additional bonds to form, creating a transition state
After the transition state when you have the enzyme product complex, and the product is released, is the enzyme regenerated?
yes
(enzyme is NOT used up in the reaction)
difference between the energy of the substrate and the products
free energy change
If a reaction releases energy, is it exergonic or endergonic?
exergonic
If a reaction absorbs energy, is it exergonic or endergonic?
endergonic
If a reaction is exergonic, is the free energy change (∆G) positive or negative?
negative
If a reaction is endergonic, is the free energy change (∆G) positive or negative?
positive
input energy needed to reach the transition state and have a chance of forming product
activation energy
What must enzymes do to reach a transition state to form the product?
**this is how enzymes speed up reactions!!
lower the activation energy
How do the enzymes lower the activation energy to reach a transition state?
bind molecules in the transition state and create a different pathway to the end product
Do low temperatures tend to increase or decrease reaction rates? Why?
decrease, reduces frequency of collision
(therefore ability of colliding molecules to reach activation energy is impaired)
Do high temperatures tend to increase or decrease reaction rates? Why?
decrease, denatures enzymes
(just a little hotter than optimum temperature will speed it up, but once the enzyme denatures, it is slowed down)
Why does pH influence enzyme-catalyzed reaction rates?
level of protonation of the enzyme's side chains has an influence on its ability to stabilize the transition state via electrostatic interactions or acid/base catalysis
(can't be too high or too low)
in Michaelis-Menton kinetics, this refers to the number of molecules of substrate converted to product in a specific amount of time
reaction rate
True or false: An enzyme can be saturated.
true
(therefore, the reaction rate depends on concentration of enzyme)
What two main factors does the reaction rate depend on?
(assuming optimal temperature and pH)
concentration of substrate, concentration of enzyme
Does increasing the substrate concentration tend to increase or decrease the reaction rate?
increase
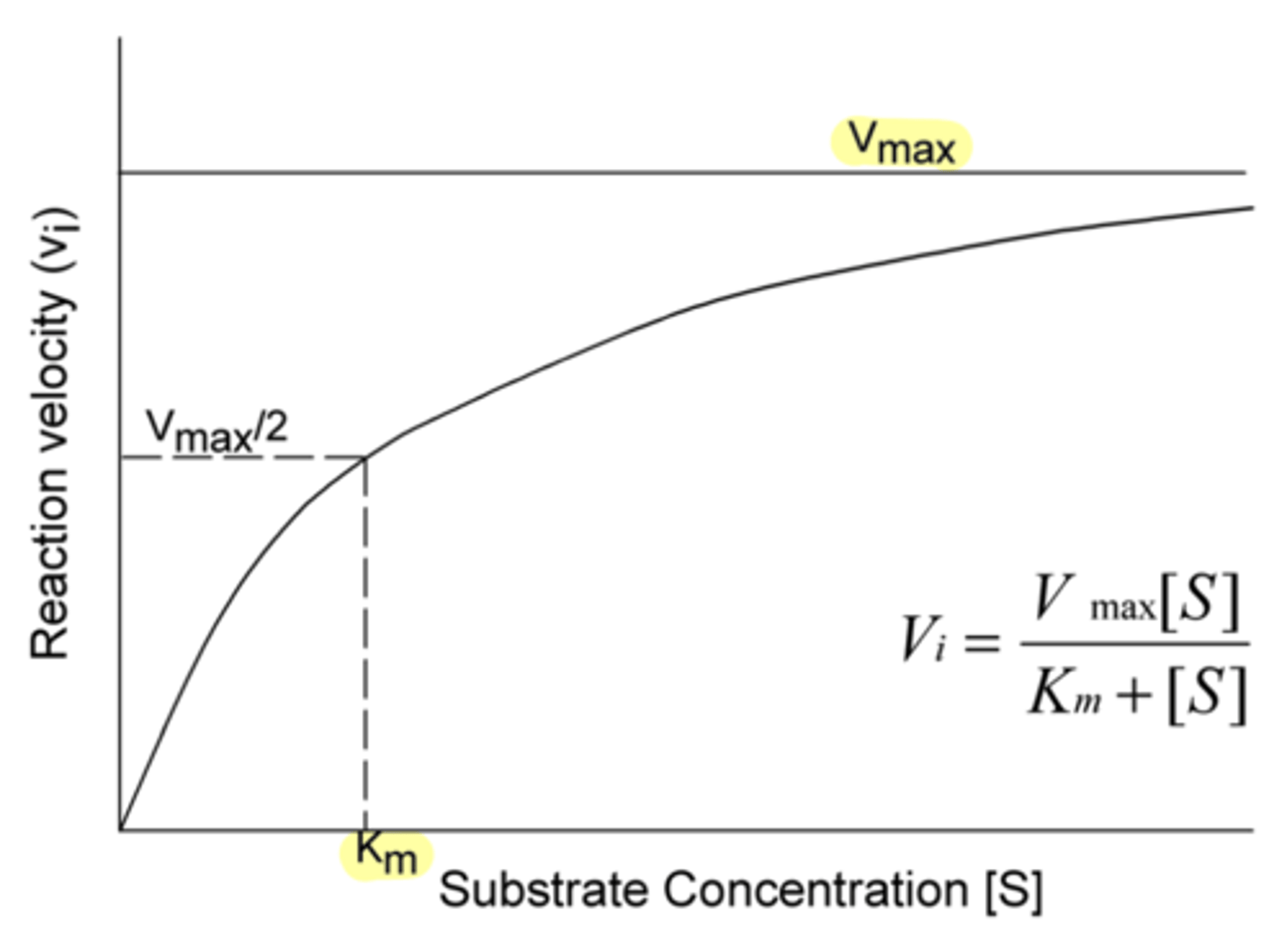
in Michaelis-Menton kinetics, this refers to the maximal rate (moles/sec) for the formation of product for a given enzyme concentration
represents the state where unlimited amounts of substrate are available and the enzyme is working as fast as it can
V-max
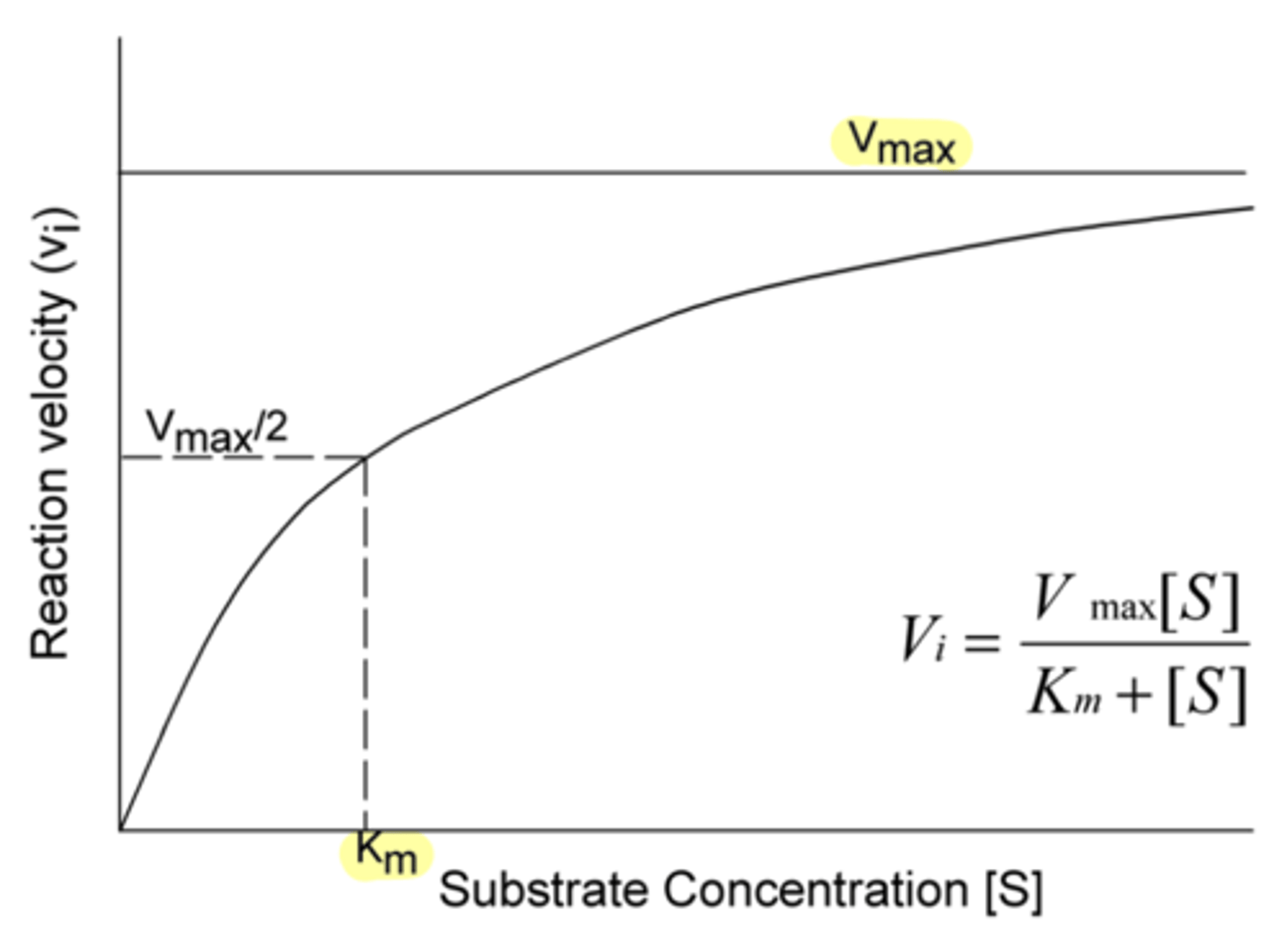
At V max, because there is an unlimited amount of substrate and the enzyme is working as fast as it can, we say that the enzyme is what?
saturated
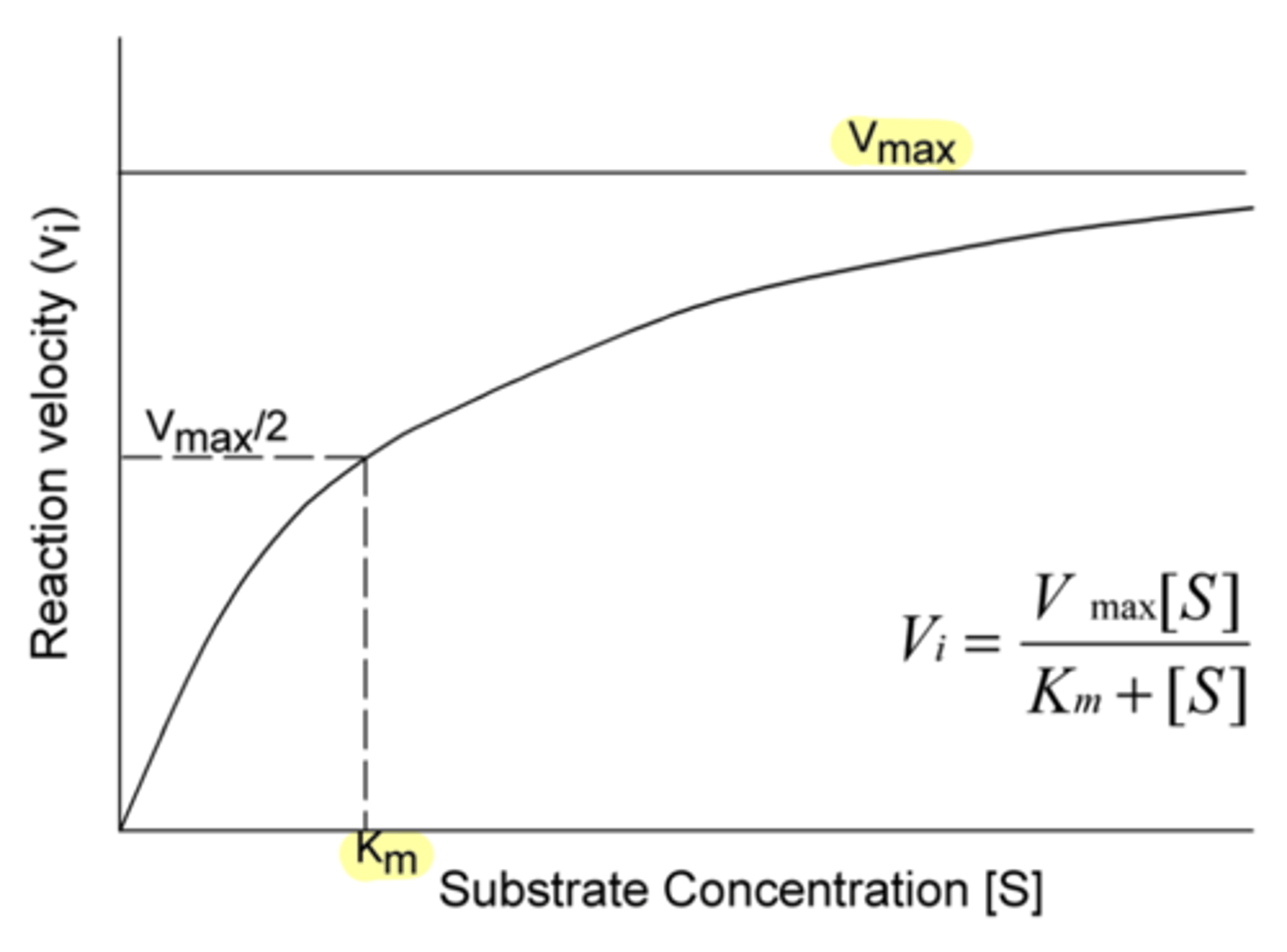
in Michaelis-Menton kinetics, this refers to the substrate concentration required to reach half of the maximum reaction rate
amount of substrate required to reach half of V max
Km
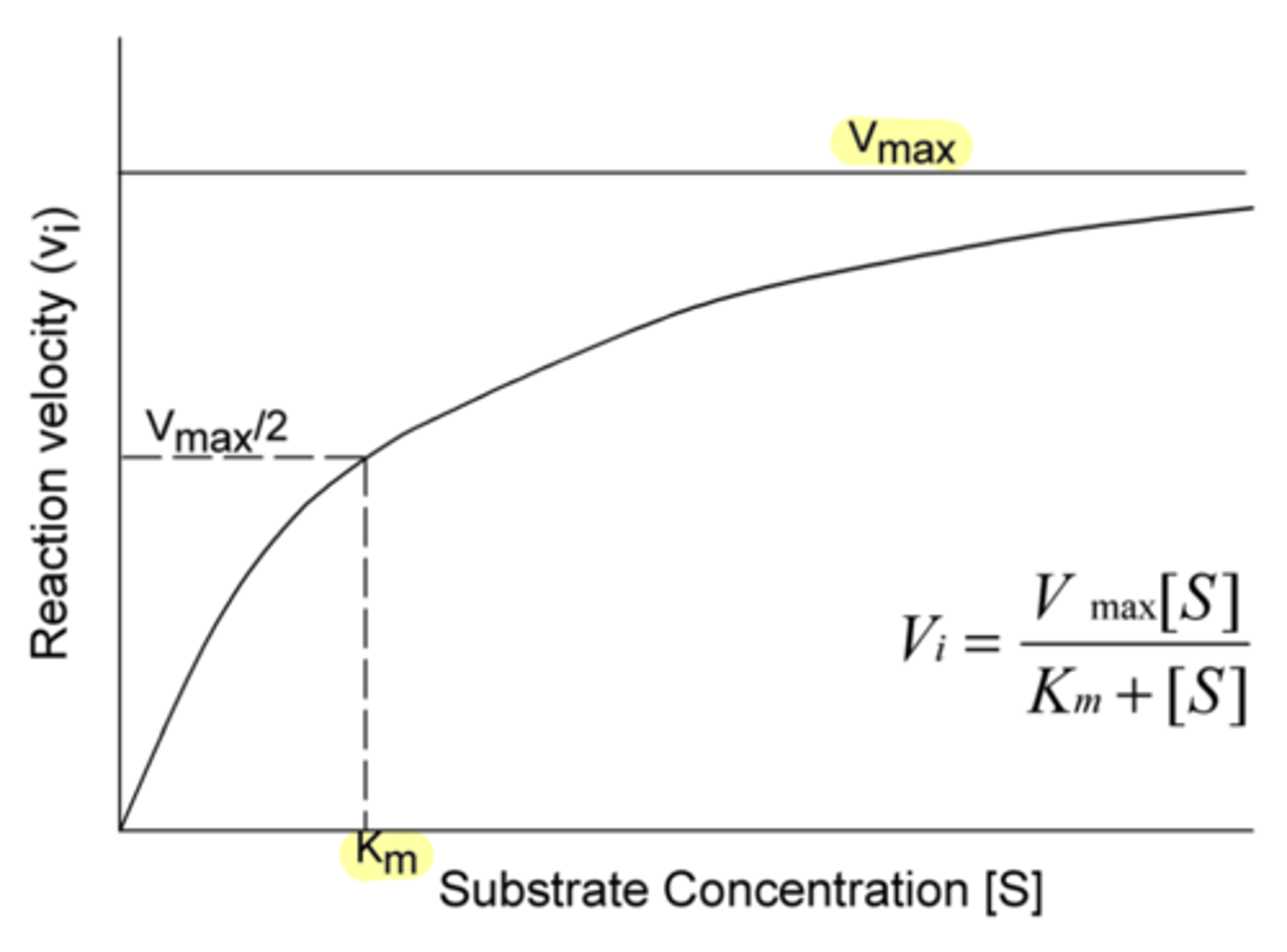
Does a large Km indicate a low or high affinity between substrate and enzyme?
low
(because more substrate is required to push the enzyme to half of V-max)
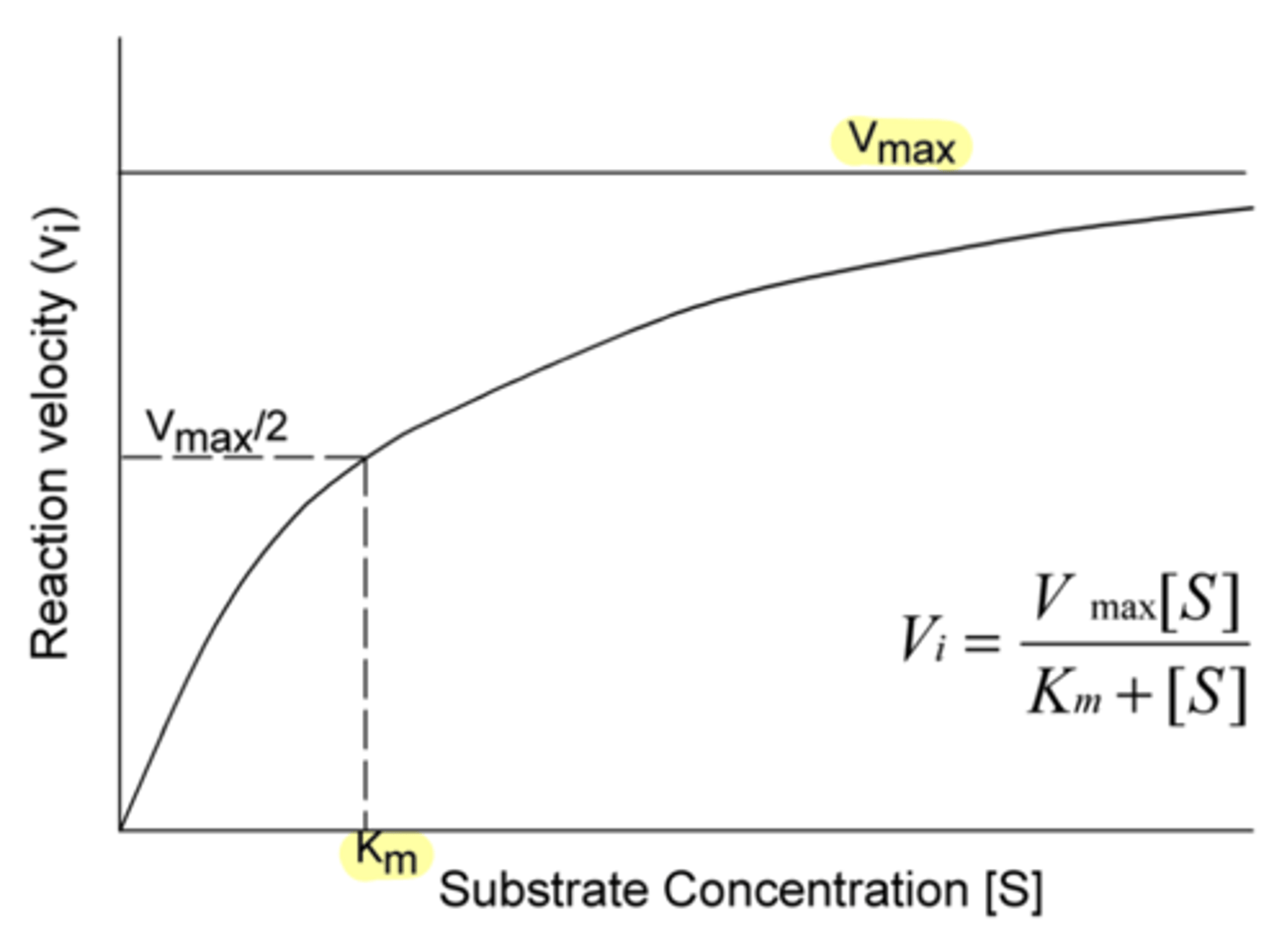
Does a small Km indicate a low or high affinity between substrate and enzyme?
high
(because less substrate is required to push the enzyme to half of V-max)
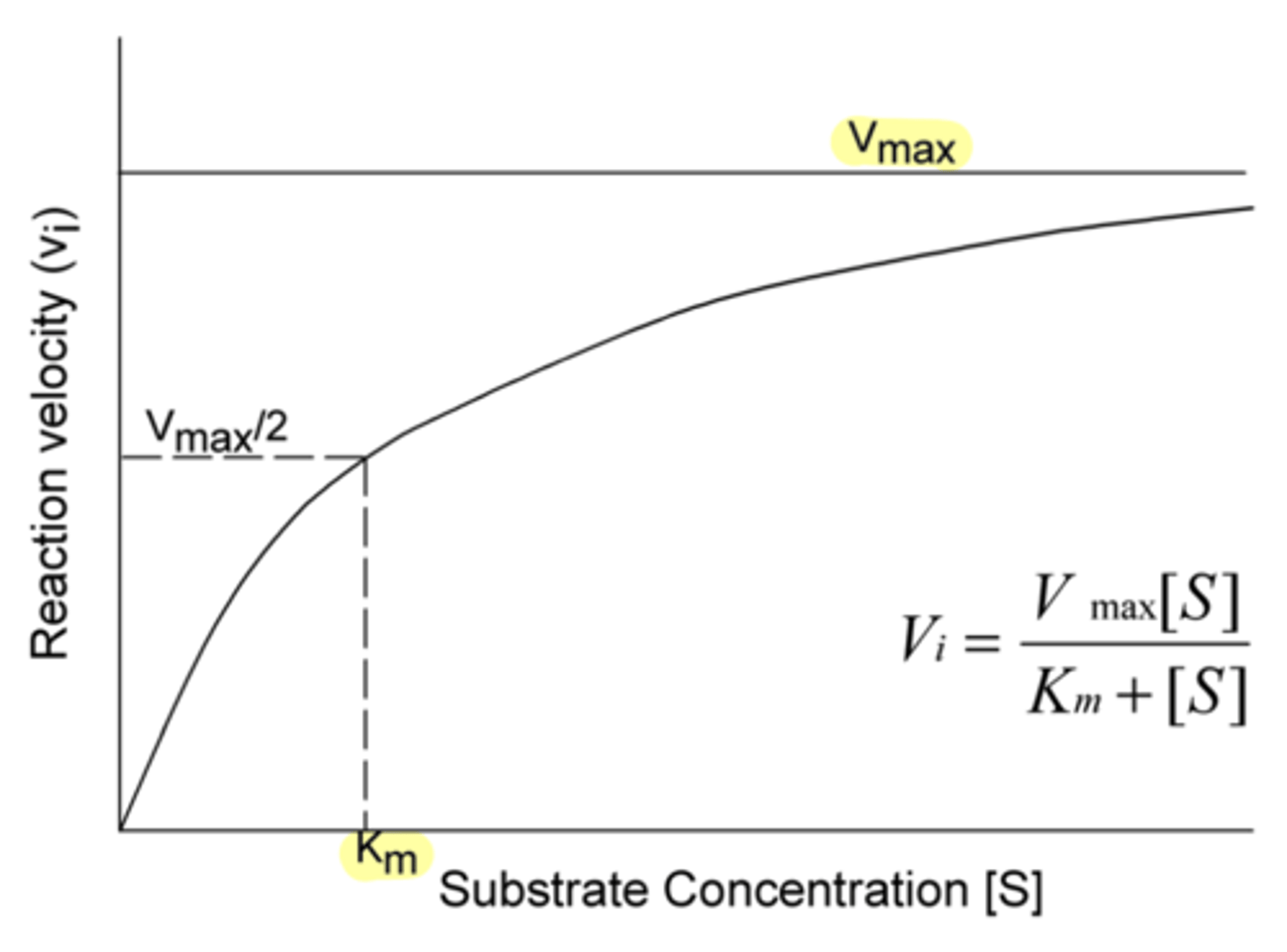
double reciprocal plot that gives us an easier way to interpret important enzyme kinetics which represent the Michaelis-Menton parameters; allows for easy determination of the kinetics
Lineweaver Burke plot
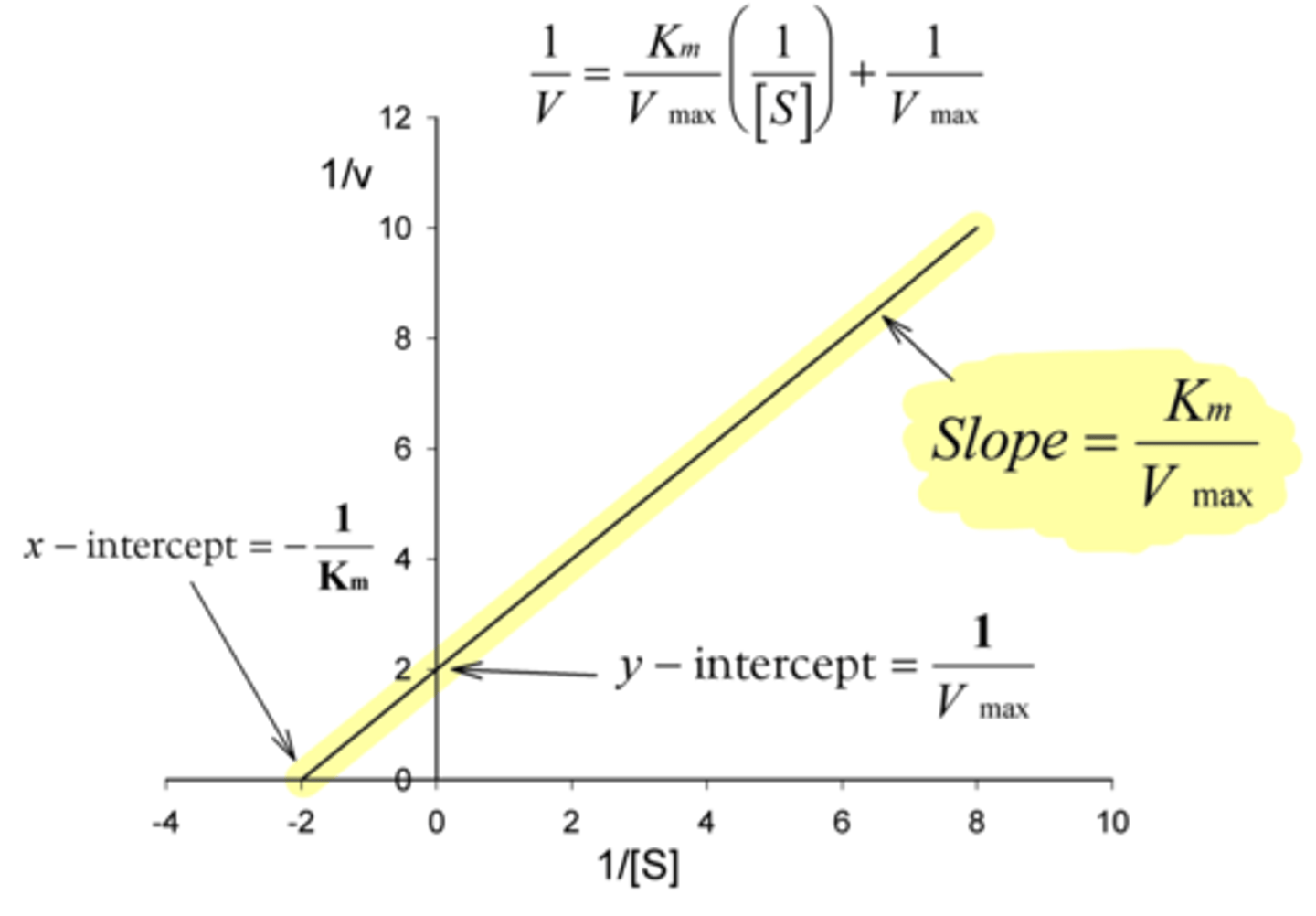
What is the slope of a Lineweaver-Burke plot?
Km/V-max
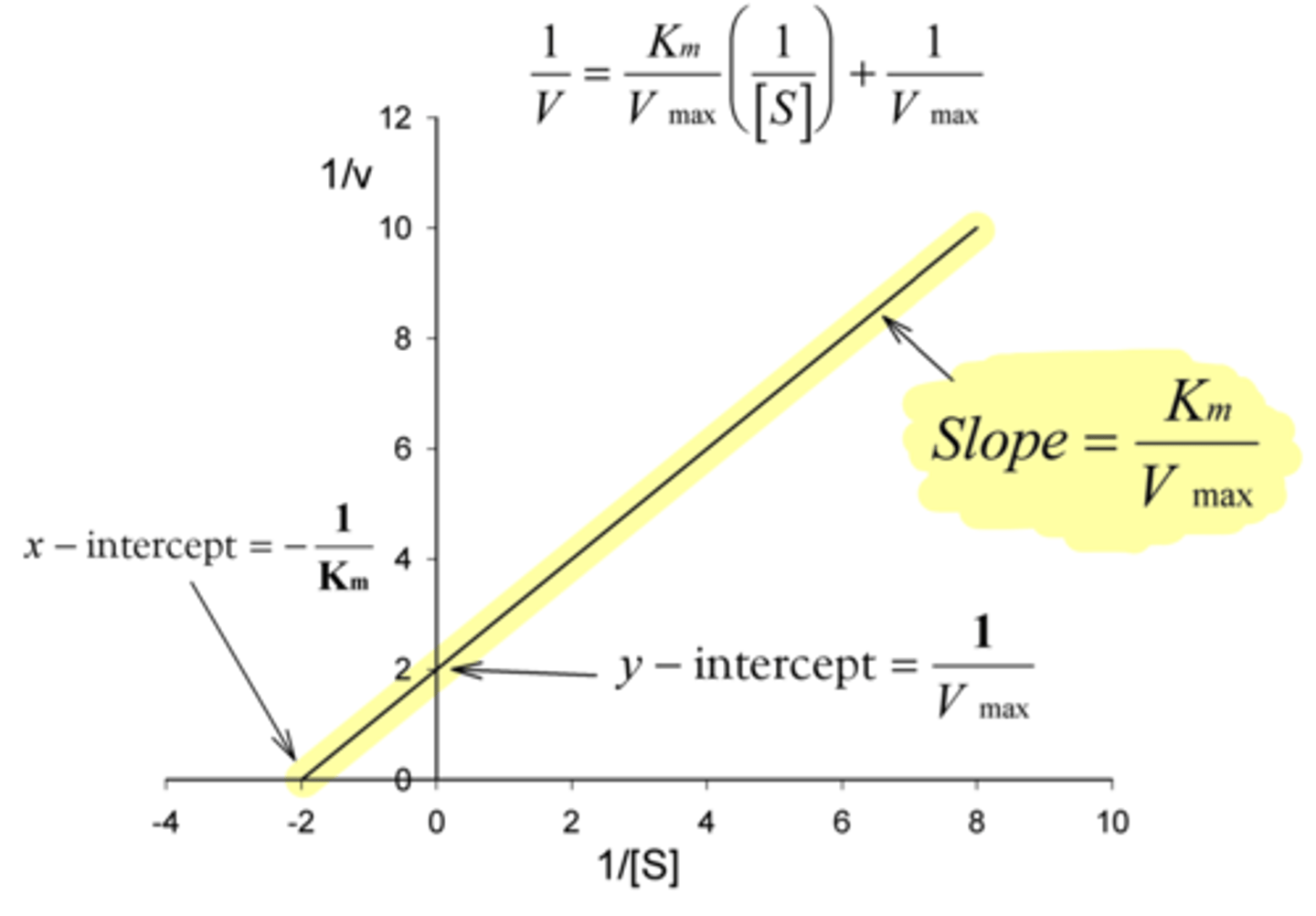
What is the x-intercept of a Lineweaver-Burke plot?
-1/Km
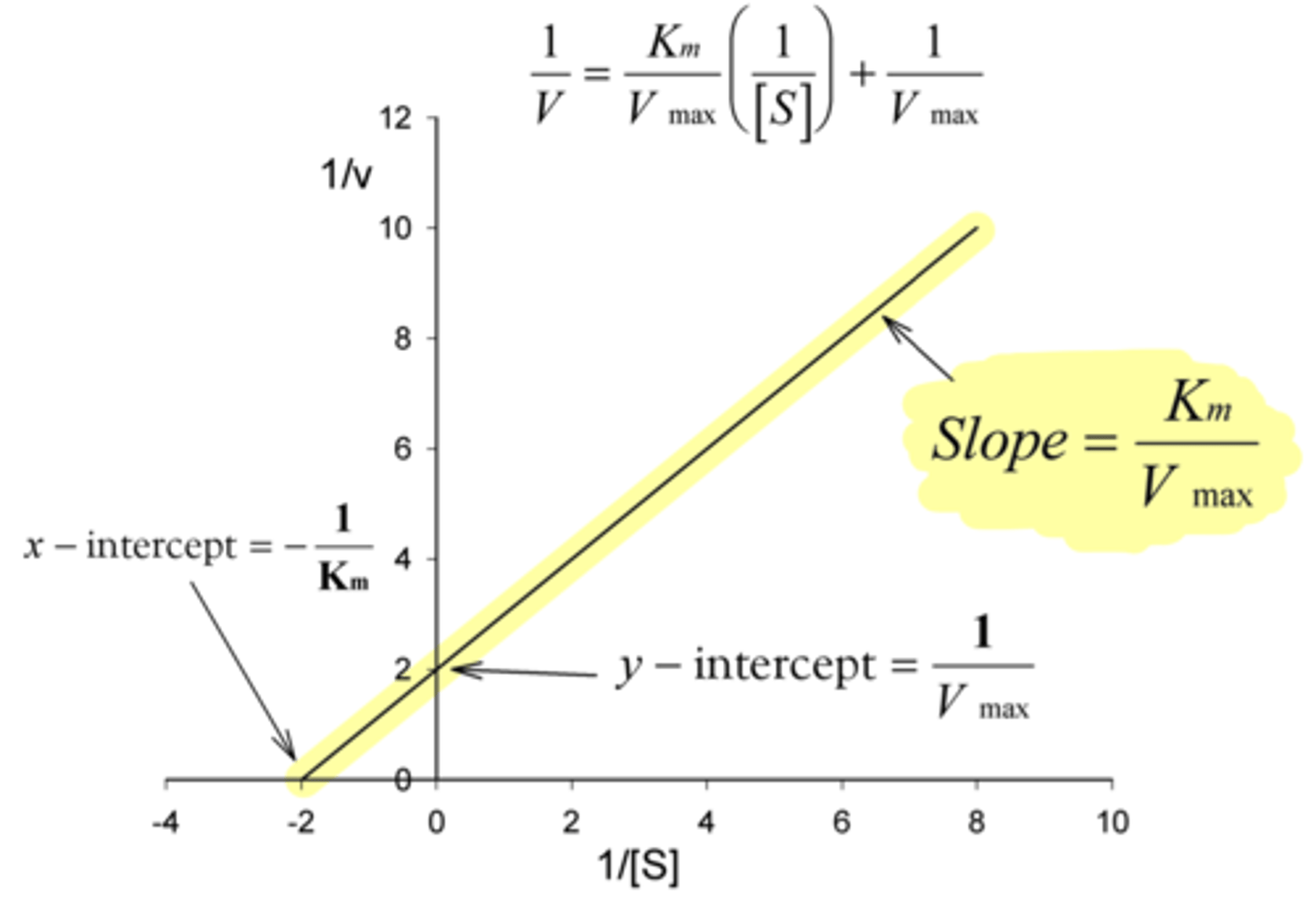
What is the y-intercept of a Lineweaver-Burke plot?
1/V-max
enzyme inhibition in which a drug non-covalently binds to the active site of an enzyme, competing with the substrate and decreasing the apparent affinity of the enzyme for the substrate
reversible competitive
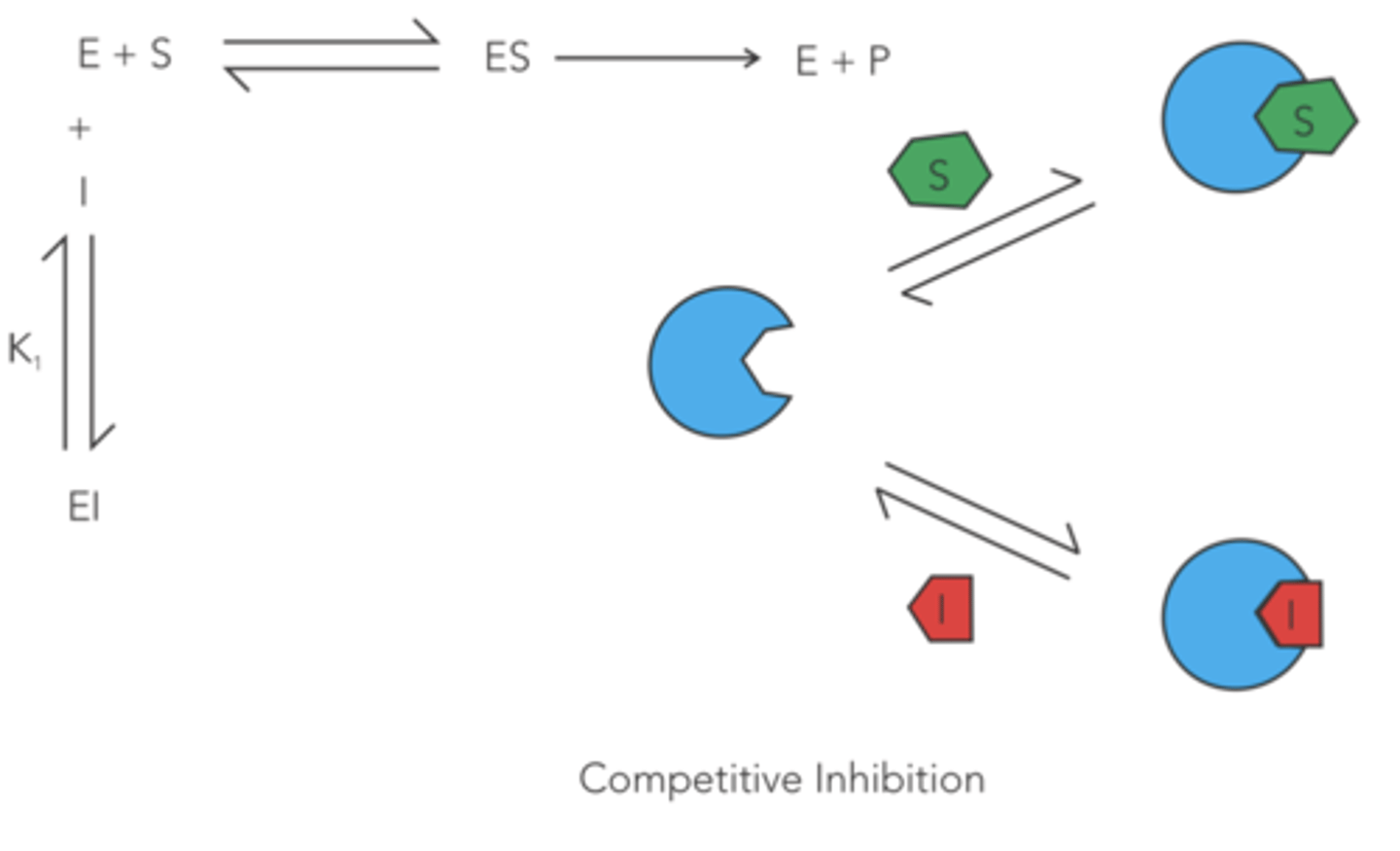
Do reversible competitive inhibitors affect Km? If so, do they increase or decrease Km?
increase
(since more substrate is needed to reach half of V max to compete with the inhibitors)
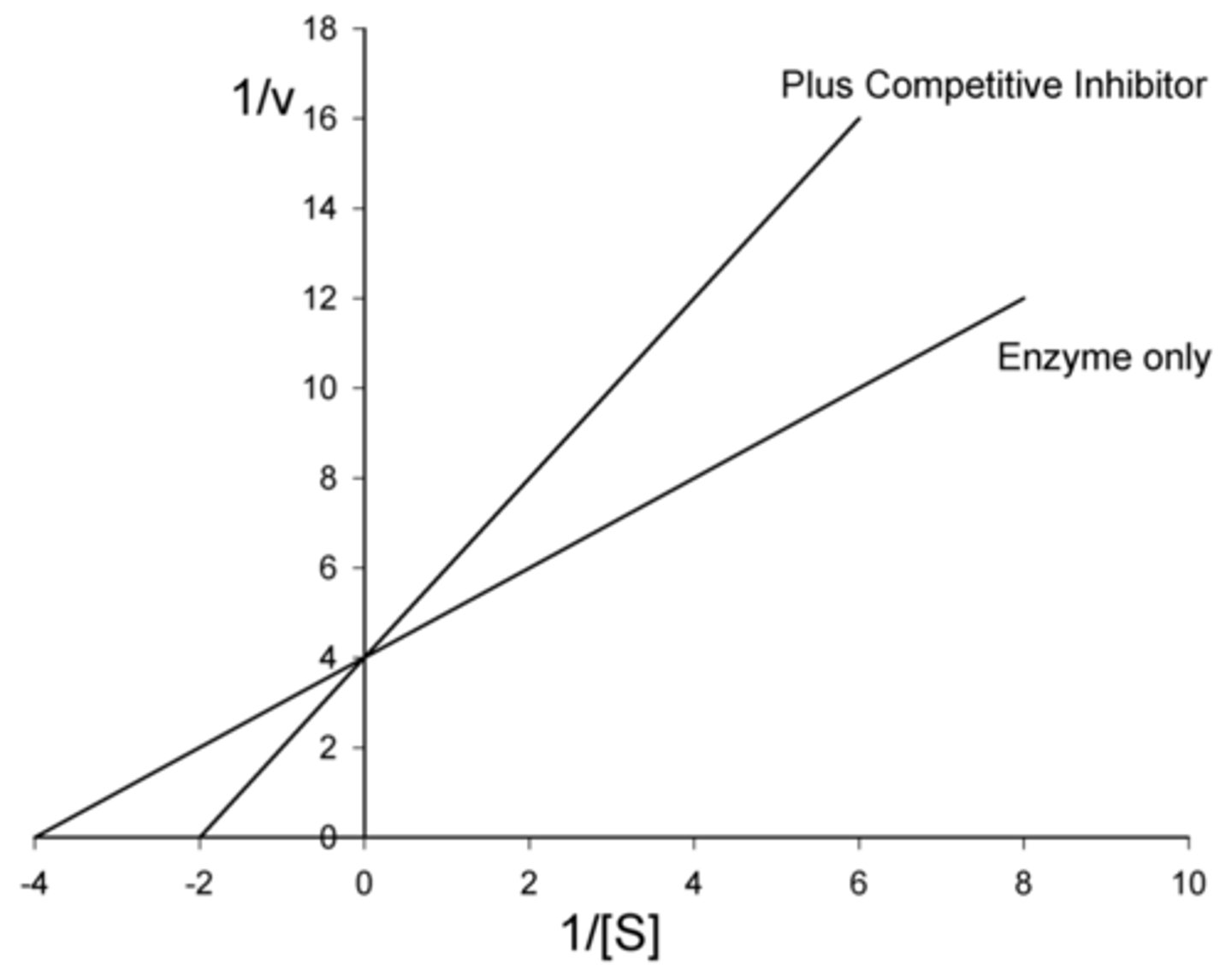
Do reversible competitive inhibitors affect V max? If so, do they increase or decrease V max?
no
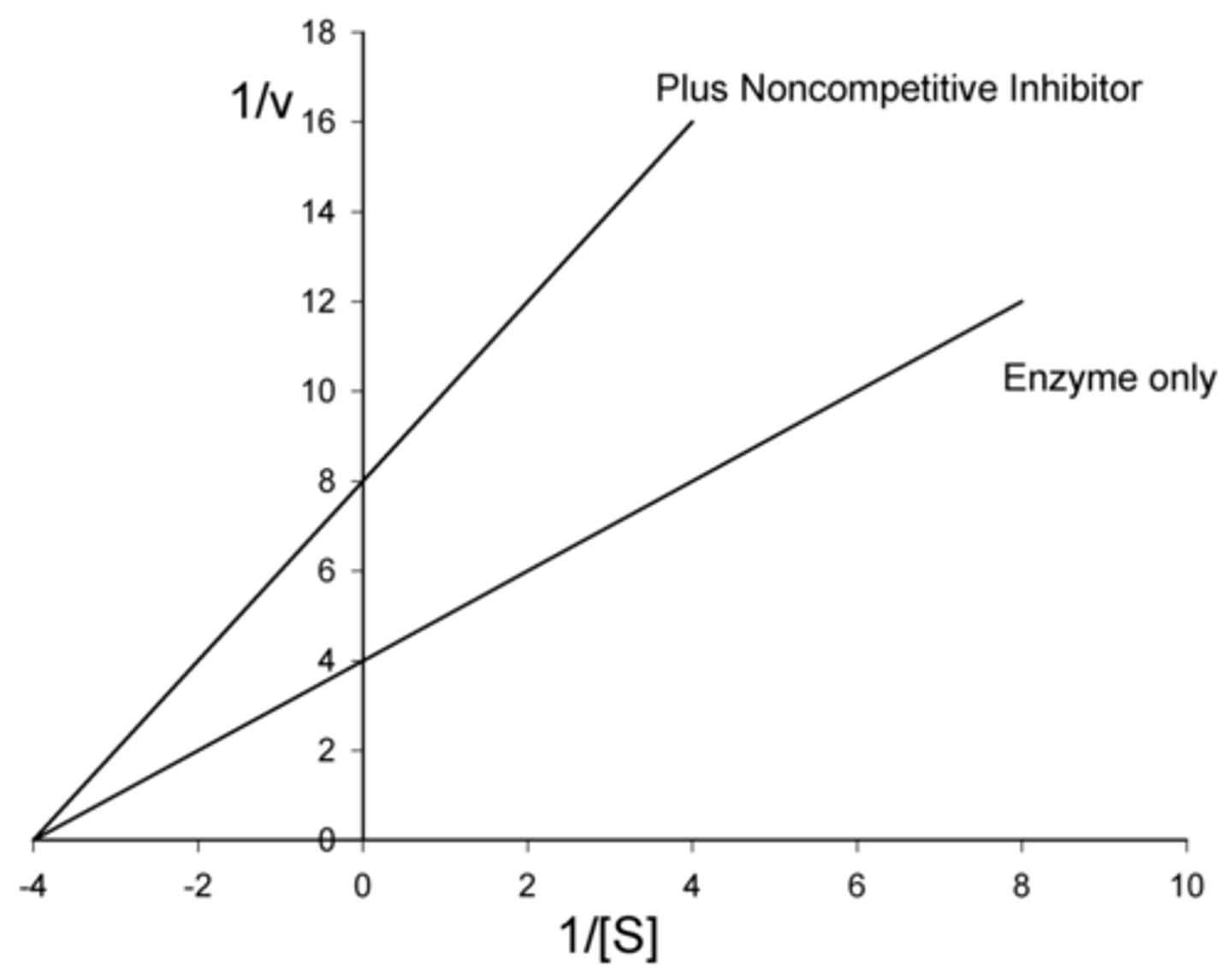
enzyme inhibition in which a drug non-covalently binds to the enzyme or the enzyme-substrate complex at a site OTHER than the active site, slowing down the rate of catalysis
reversible noncompetitive
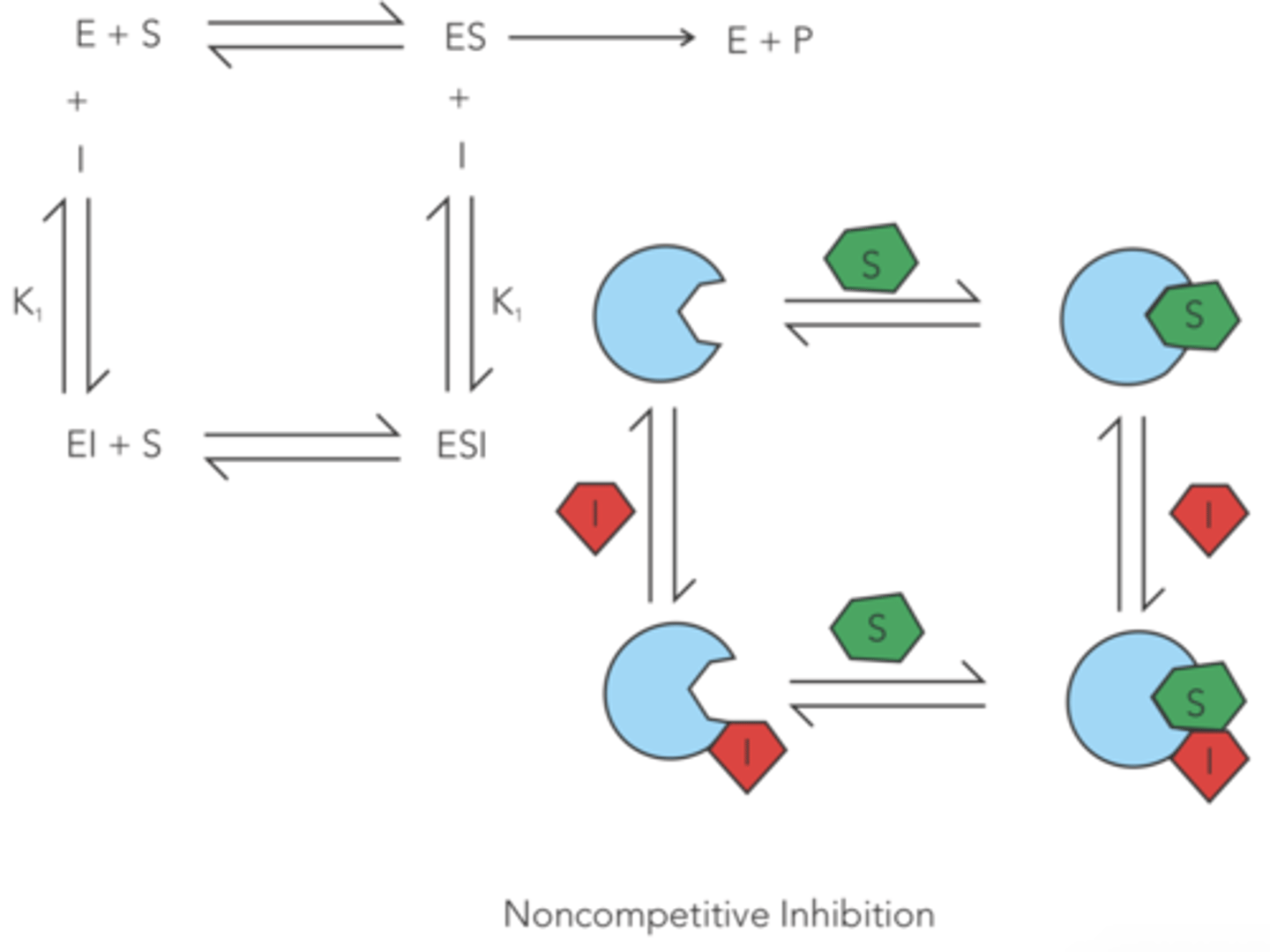
Do reversible noncompetitive inhibitors affect V max? If so, do they increase or decrease V max?
decrease
(since product cannot be formed as long as the inhibitor is bound to the enzyme)
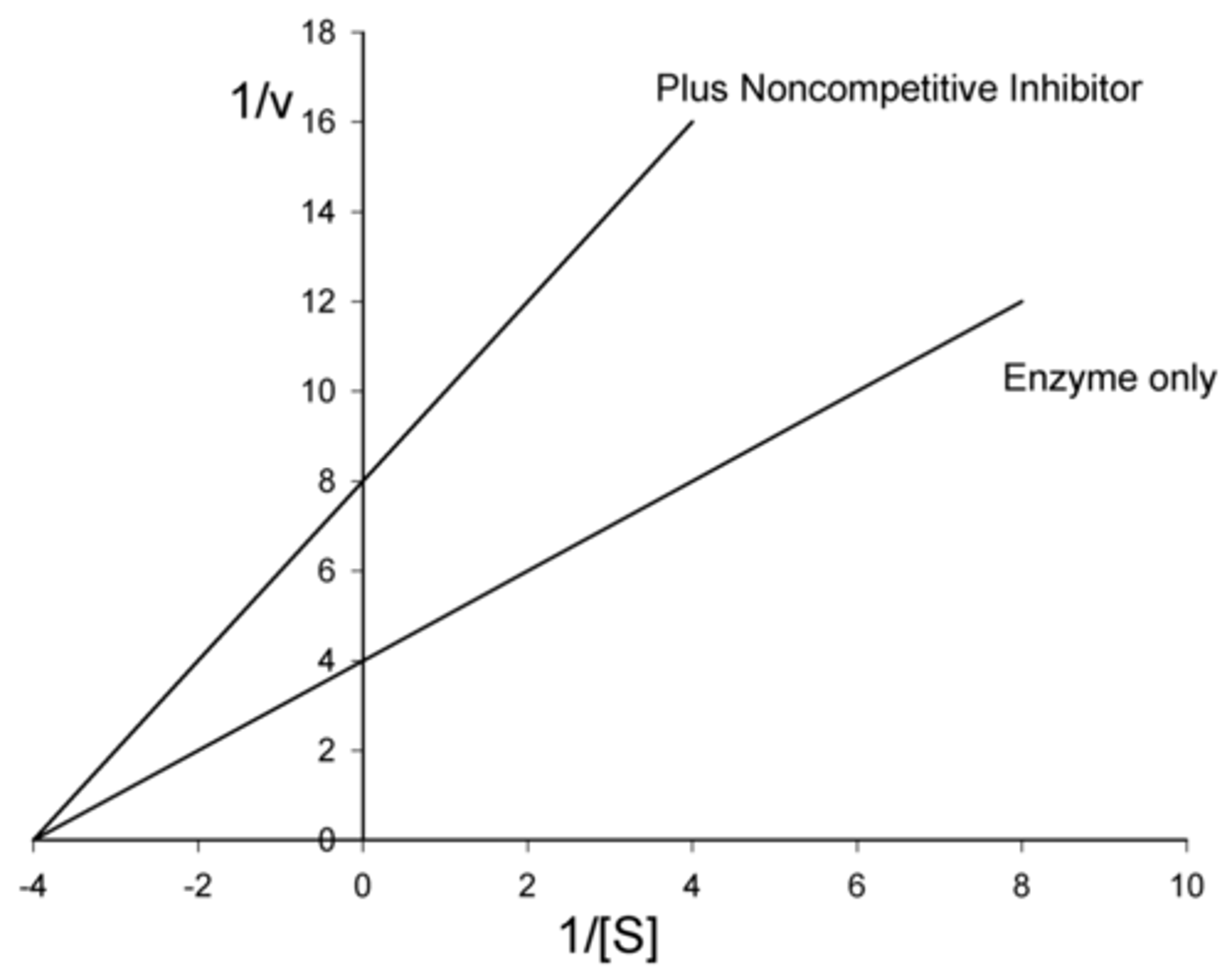
Do reversible noncompetitive inhibitors affect Km? If so, do they increase or decrease Km?
no
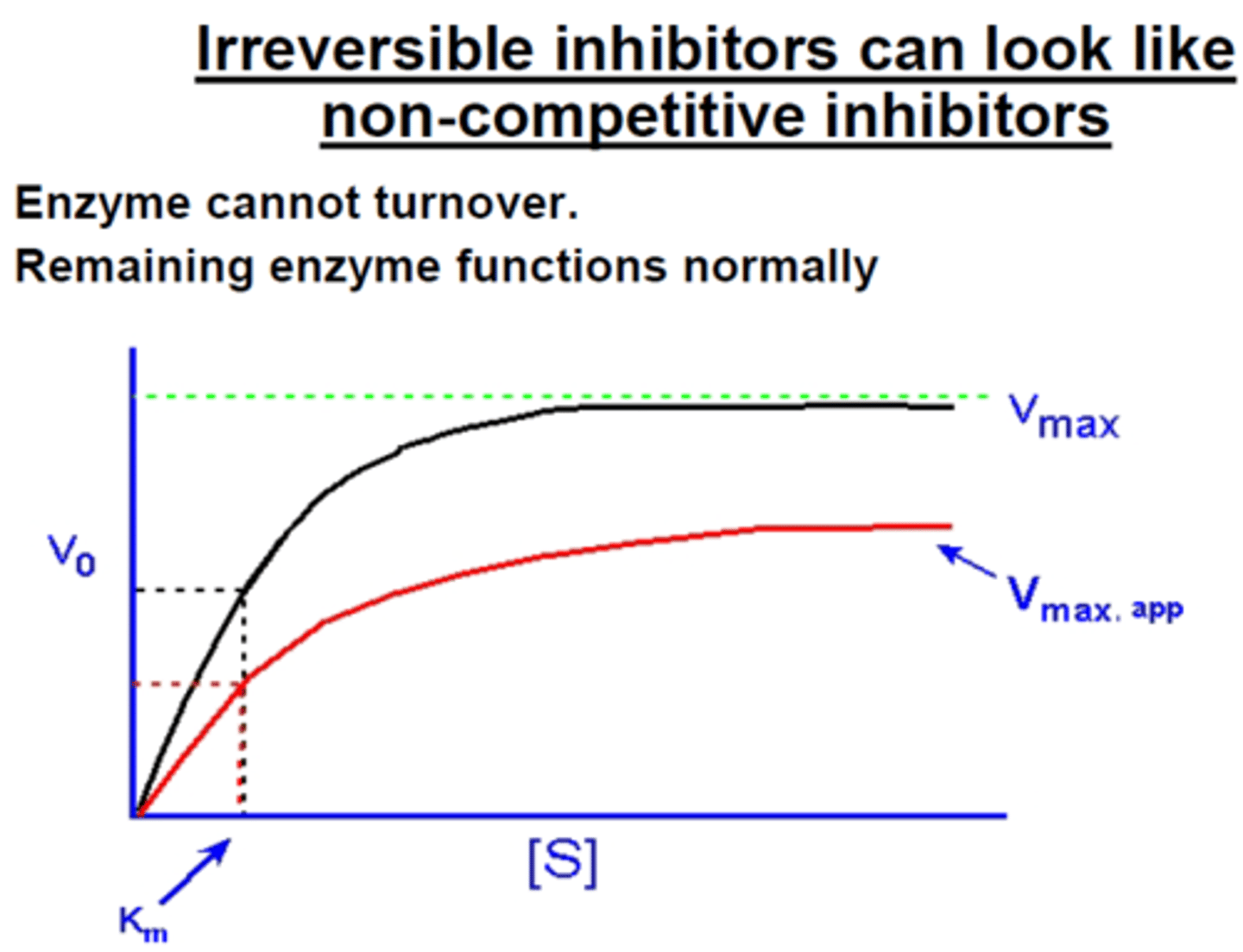
enzyme inhibition in which a drug covalently modifies the enzyme structure, resulting in activation of the enzyme
**lowers the overall reaction rate in the body since the enzyme is basically lost
irreversible (competitive)
Do irreversible inhibitors affect V max? If so, do they increase or decrease V max?
decrease
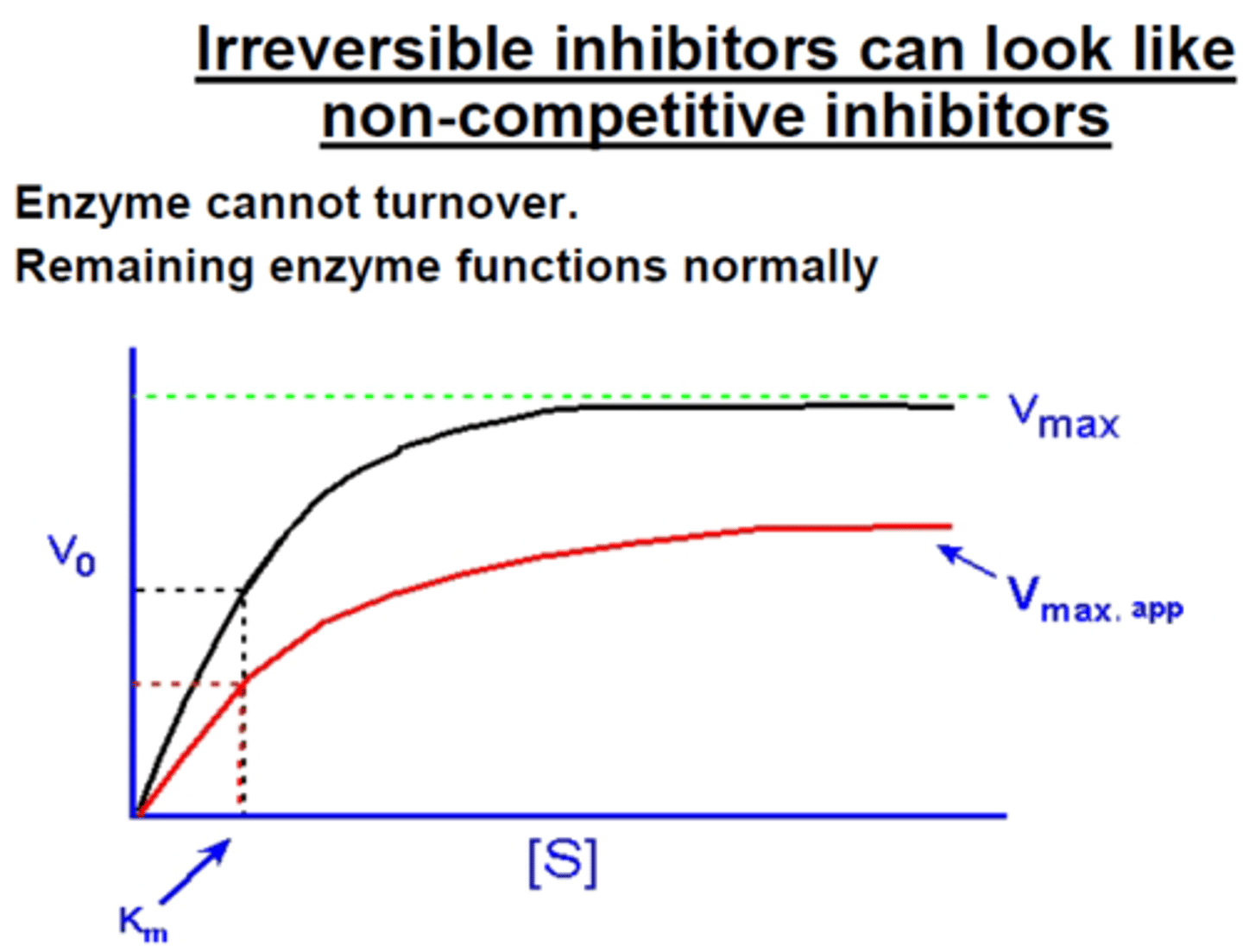
Do irreversible inhibitors affect Km? If so, do they increase or decrease Km?
no
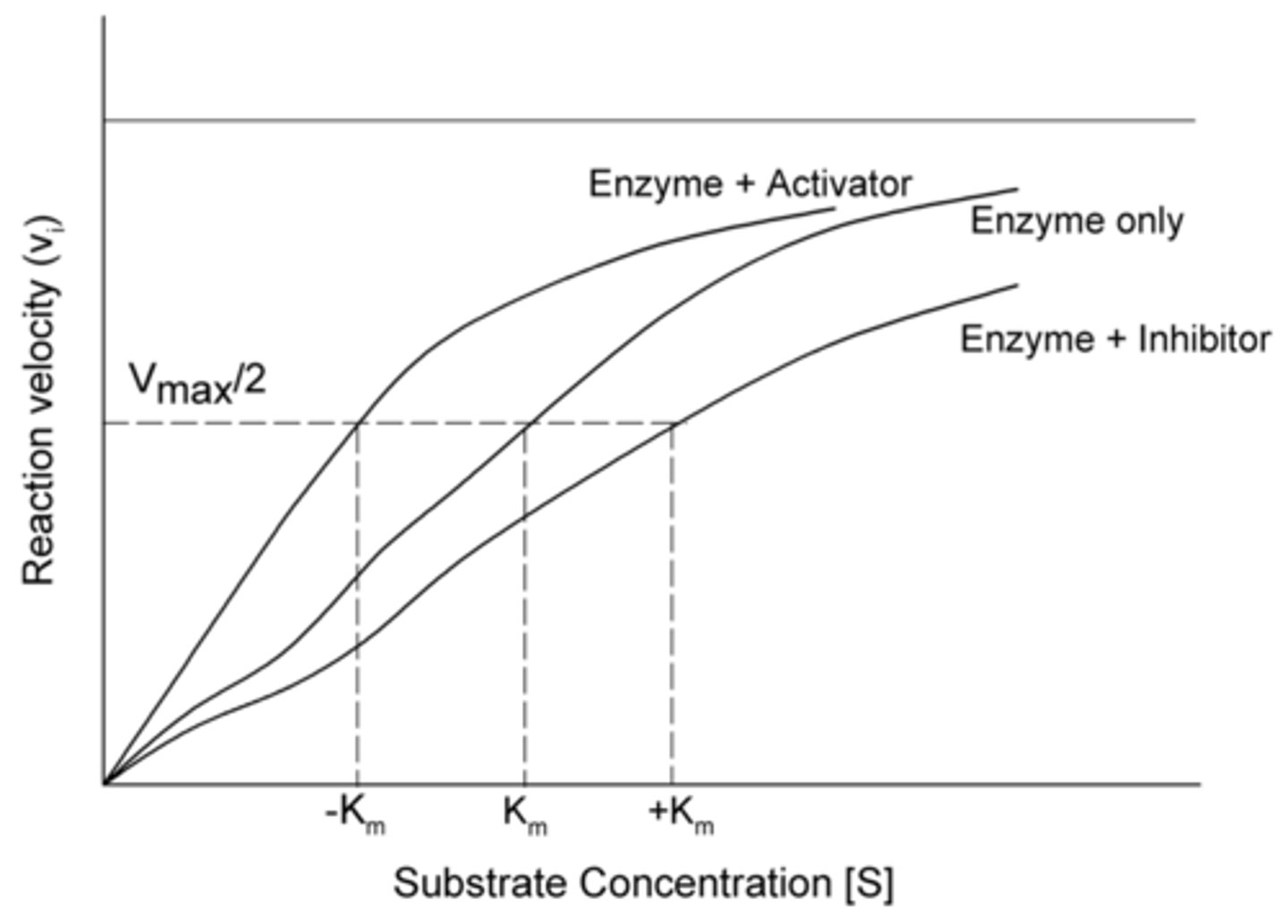
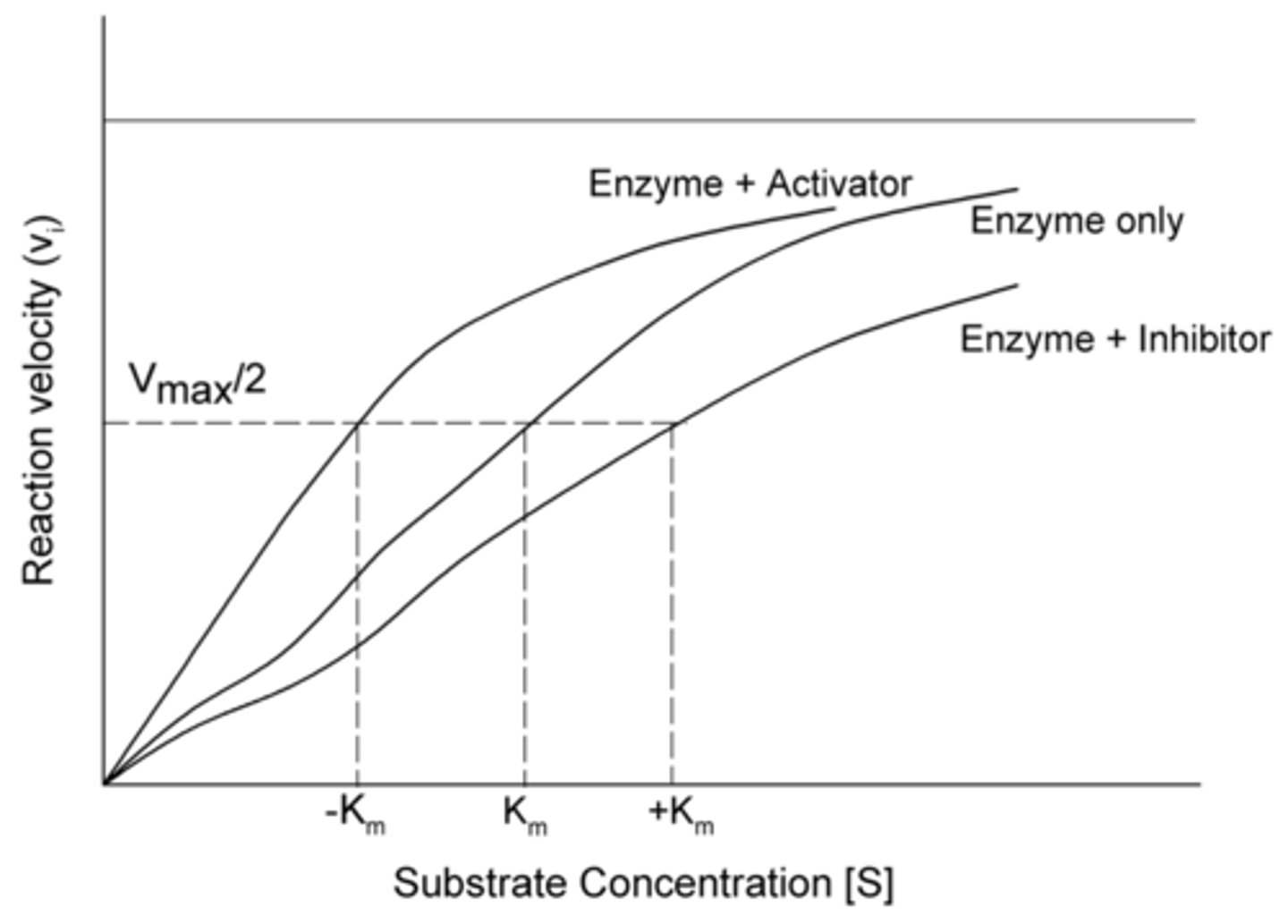
When allosteric activators bind to an enzyme, it changes the catalytic binding site so that it increases or decreases the affinity for the substrate? Is this an increase or decrease in Km?
increases affinity, decreases Km
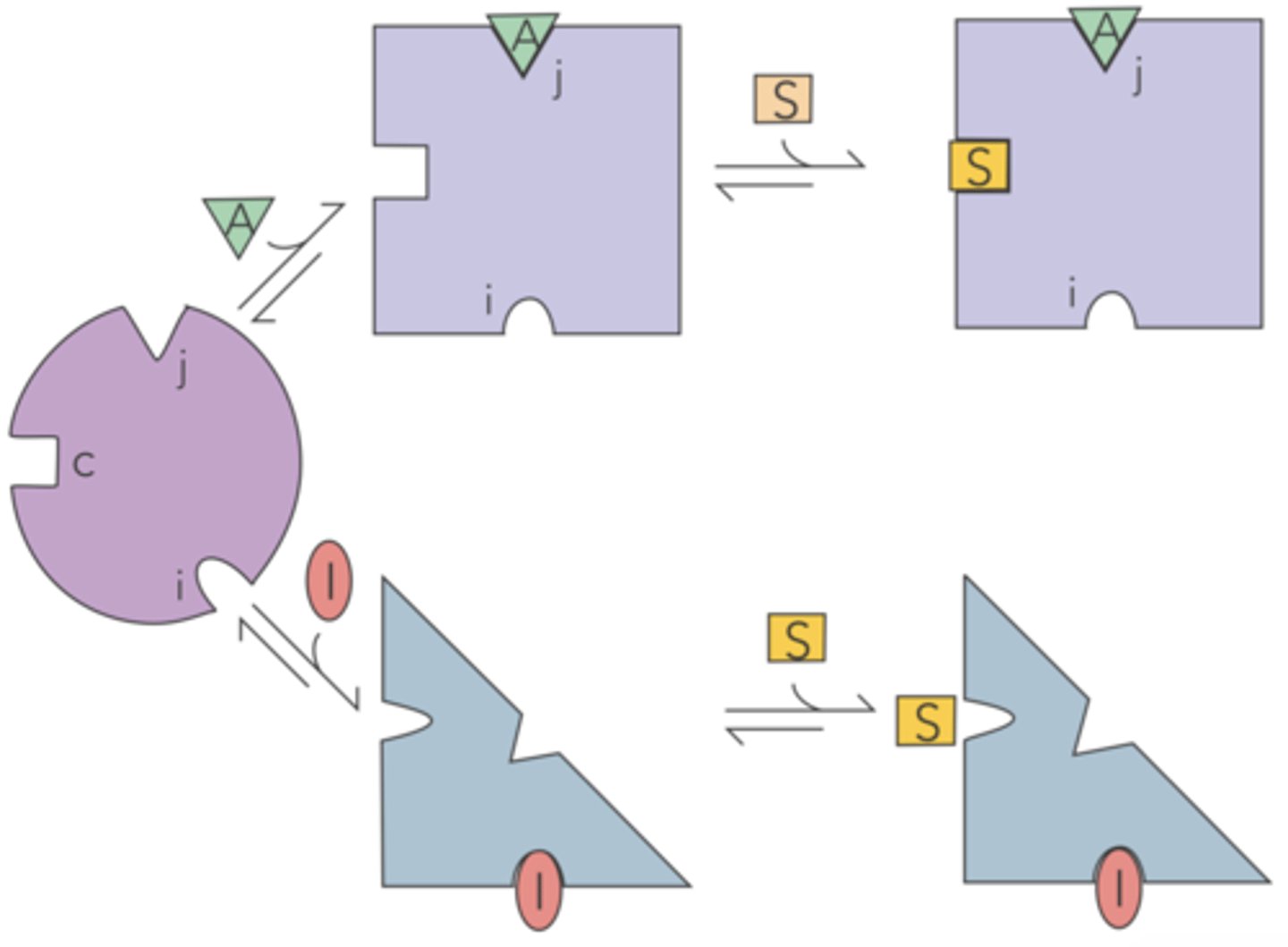
refers to enzymes that contain three specific binding sites: catalytic, activator, and inhibitor
something affects this enzyme outside of the active site
allosteric
When allosteric inhibitors bind to an enzyme, it changes the catalytic binding site so that it increases or decreases the affinity for the substrate? Is this an increase or decrease in Km?
decreases affinity, increases Km
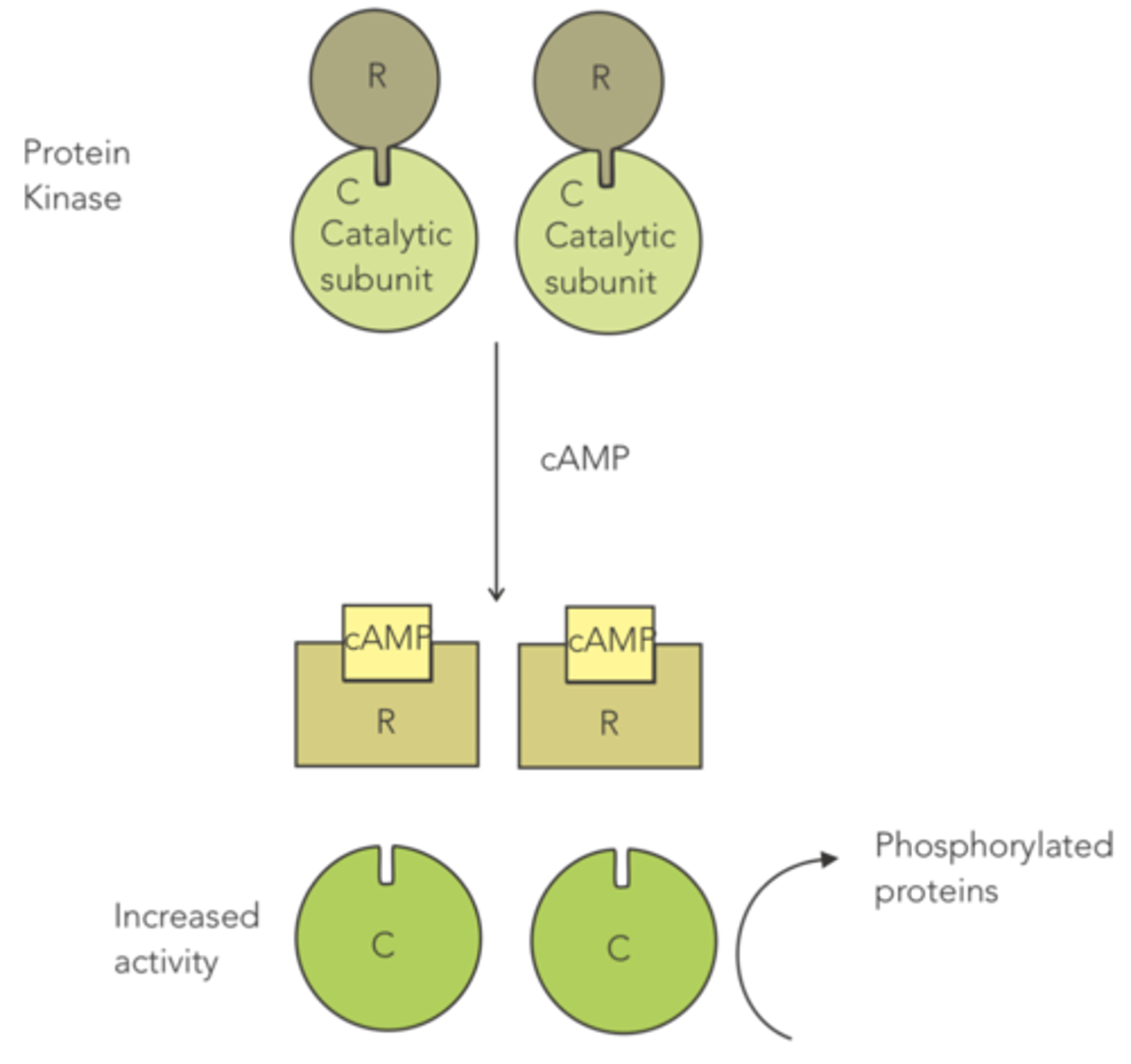
enzyme that contains a regulatory subunit that acts as an allosteric inhibitor, since it binds to the catalytic subunit to prevent high levels of activity
when the cell wants higher levels of protein kinase activity, cAMP is released and causes the regulatory subunit to unbind, so that the enzyme can phosphorylate proteins
protein kinase
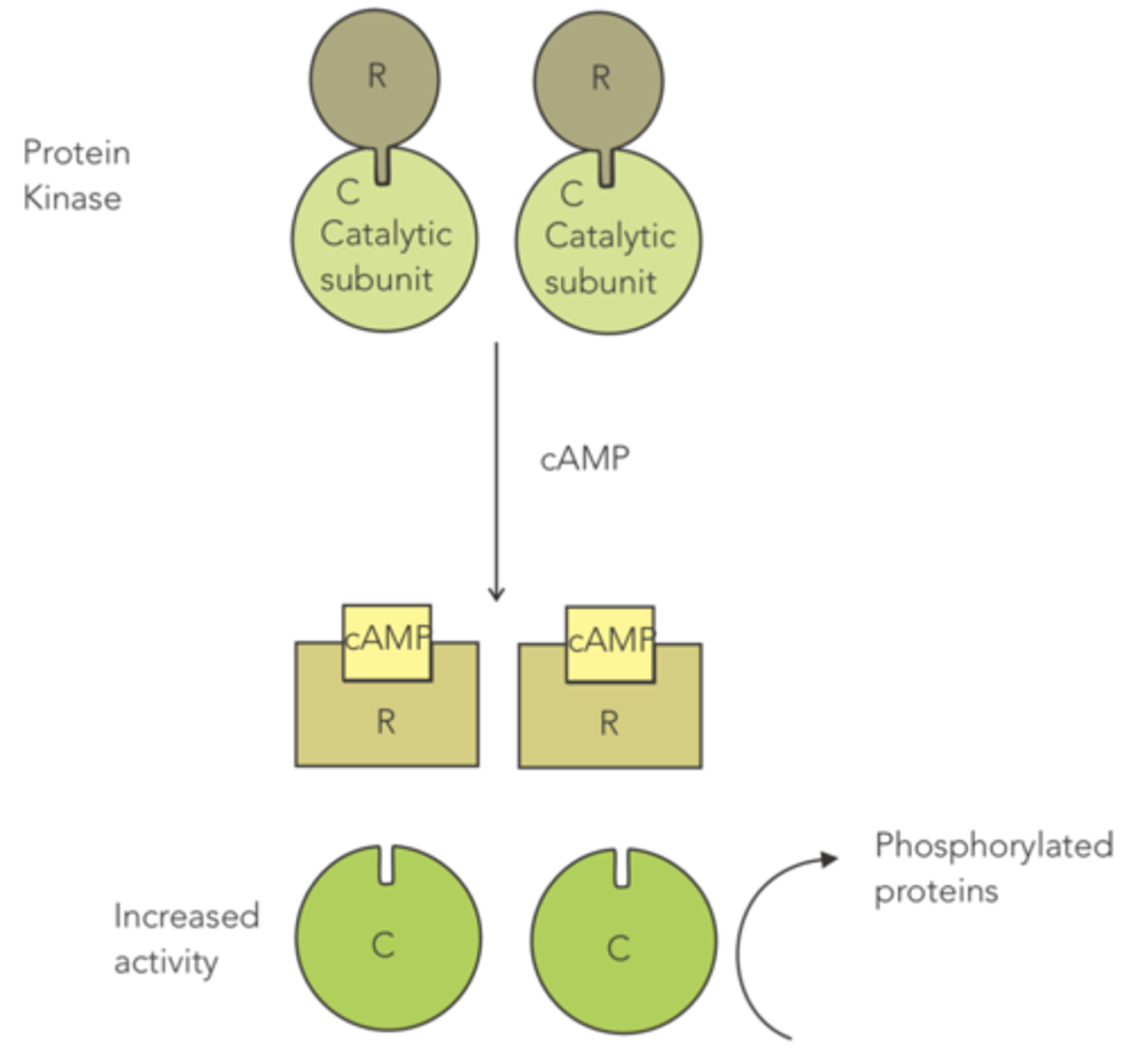
How is protein kinase an example of an allosteric enzyme?
it contains a regulatory subunit that serves as an allosteric inhibitor, since it binds to the catalytic subunit to prevent high levels of activity
(when the cell wants higher levels of protein kinase activity, cAMP is released, causing the regulatory subunit to unbind)
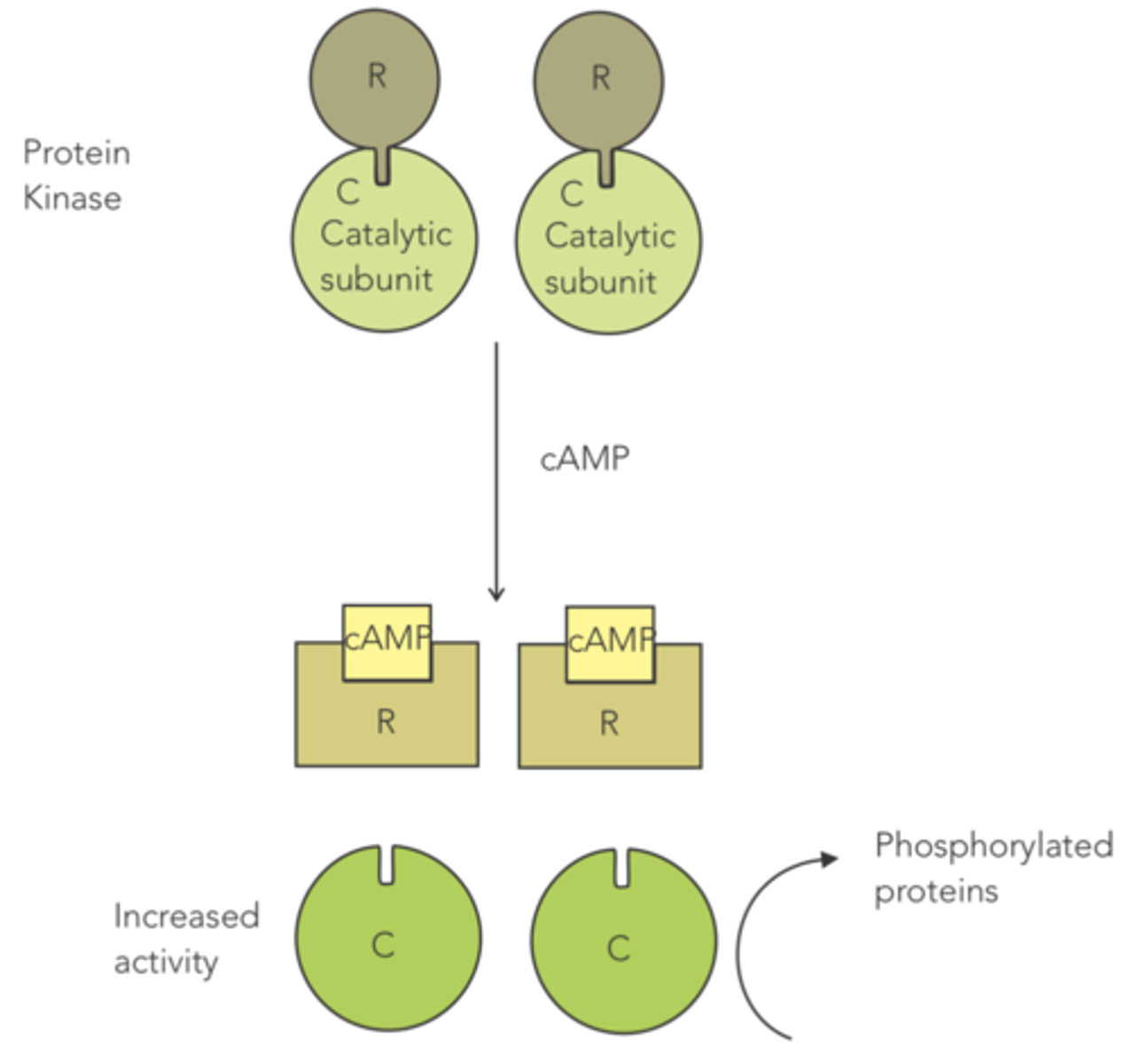
Protein kinase contains a regulatory subunit that acts as an allosteric inhibitor, since it binds to the catalytic subunit to prevent high levels of activity.
When the cell wants higher levels of protein kinase activity, what is released that causes the regulatory subunit to unbind?
cAMP
Precursor cleavage of zymogens or pro-enzymes leads to a cascade activation, where what happens?
**used during digestion and blood clotting
the products of each reaction enable the next reaction to proceed
occurs when the concentration of products of a reaction controls the reaction rate by affecting an upstream step, usually the rate limiting step or first committed step
feedback regulation
Feedback regulation occurs when the concentration of products of a reaction controls the reaction rate by affecting an upstream step, usually the what?
rate limiting step
(or first committed step)
refers to when the products of a reaction inhibit the reaction that formed it
product inhibition
refers to when the products of a reaction inhibit a reaction that occurred several steps back in the cascade (usually the rate-limiting step or first committed step)
end product inhibition
type of feedback that occurs when high product concentrations inhibit further synthesis of the product, thereby keeping the concentration of the product in a tightly controlled range
negative
type of feedback that occurs when high product concentrations increase further synthesis of the product, thereby increasing both the rate of product formation and the amount of product until the substrate is exhausted
positive
Are most physiological systems under positive or negative feedback control?
negative
What does lysozyme in the aqueous layer of tears do?
destroy gram-positive bacteria
What is the structure of the cell wall of bacteria?
alternating NAG and NAM residues joined by a beta 1-4 linkage with no outer lipid membrane
What are the two alternating residues that make up a bacterial cell wall?
NAM, NAG
How does lysozyme disrupt the bacterial cell wall?
hydrolyzes glycosidic bond between C-1 of NAM and C-4 of NAG
(causes bacterial cell to swell and burst)
Which of the two alternating residues that makes up a bacterial cell wall has the bulkier side chain: NAM or NAG?
NAM
(bulky side chains of NAMs allow them to line up against the lysozyme for hydrolysis)
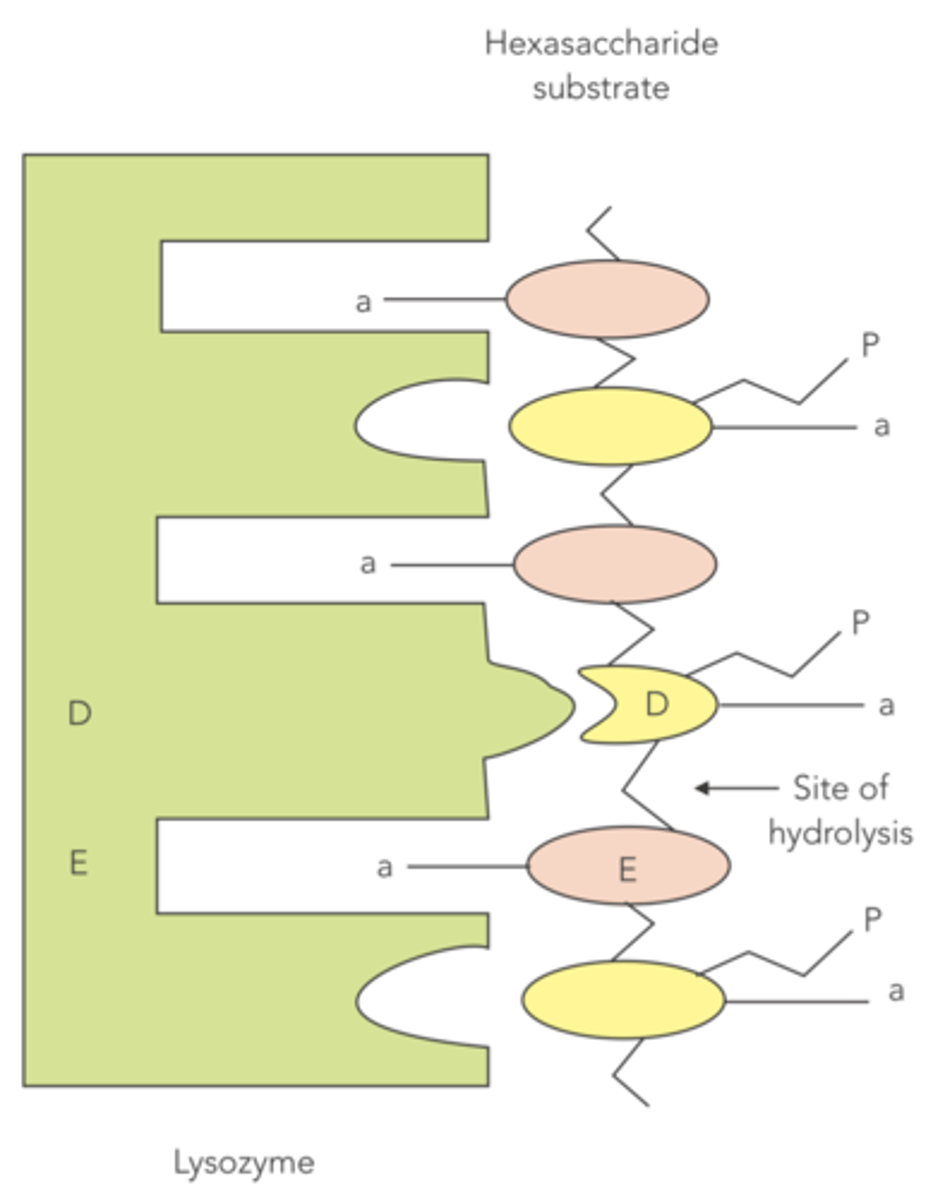
NAG and NAM align at the active site of lysozyme such that the bulkier side chains of NAM help orient the sugar groups in the active site by what?
steric hindrance
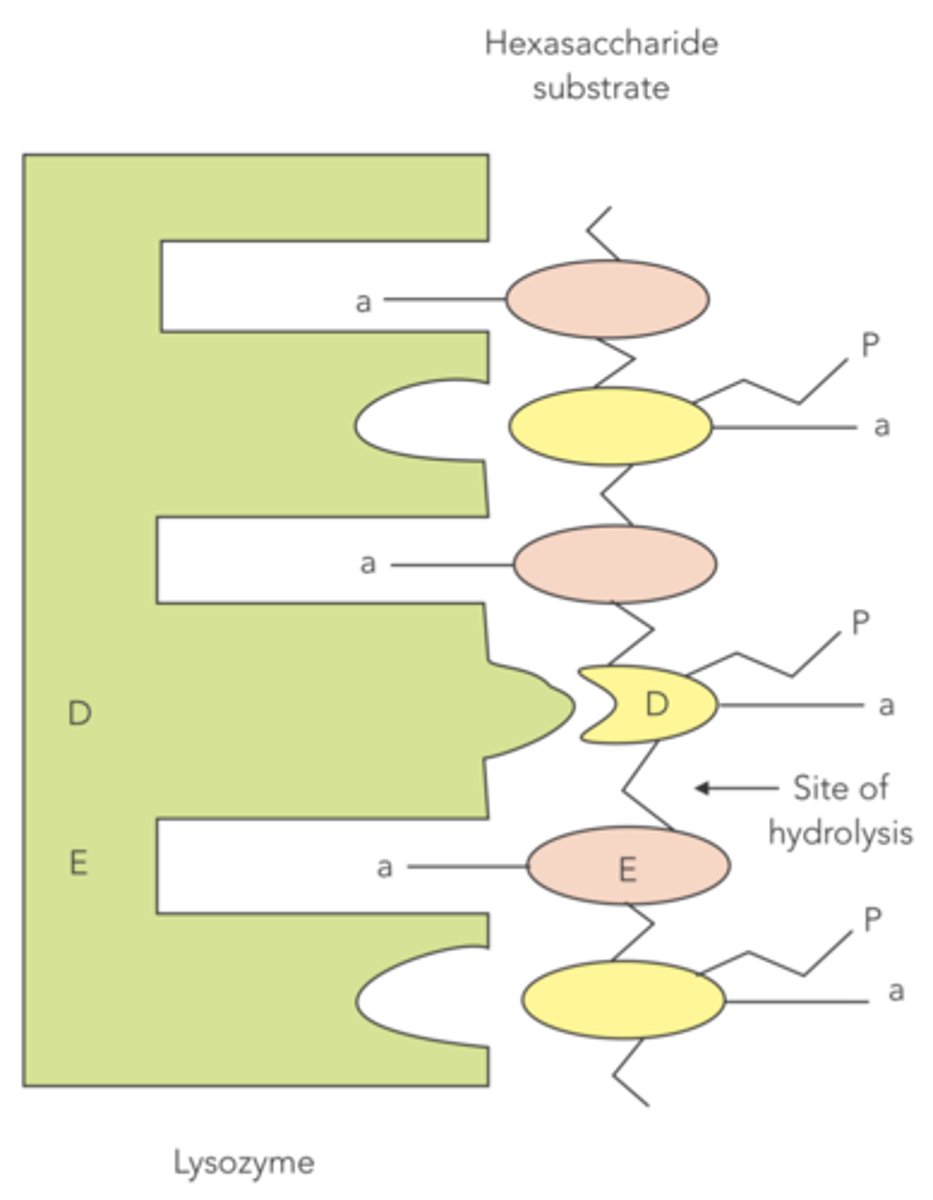
The mechanism of catalysis of the lysozyme destroying certain gram-positive bacteria is a combination of what three things?
substrate strain, acid-base catalysis by glutamate, transition state stabilization by aspartate
How do the bulky side chains of NAM on bacterial cell walls cause substrate strain?
aligns the cell wall into the active site of lysozyme, which forces the sugar structure into an unstable conformation and puts strain on the glycosidic bond
What amino acid is responsible for acid-base catalysis in the reaction between lysozyme and bacterial cell walls? What does it do?
glutamate, donates a proton to NAG to break the glycosidic bond
What amino acid is responsible for transition state stabilization in the reaction between lysozyme and bacterial cell walls? What does it do?
aspartate, interacts with the cation on the NAM
After lysozyme has hydrolyzed the cell wall of gram-positive bacteria, the newly disrupted peptidoglycan leaves the active site, and what regenerates the enzyme?
water