BIOCHEM - All 20 Amino Acids, Protease, Types of Globins and Proteins
1/61
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
62 Terms
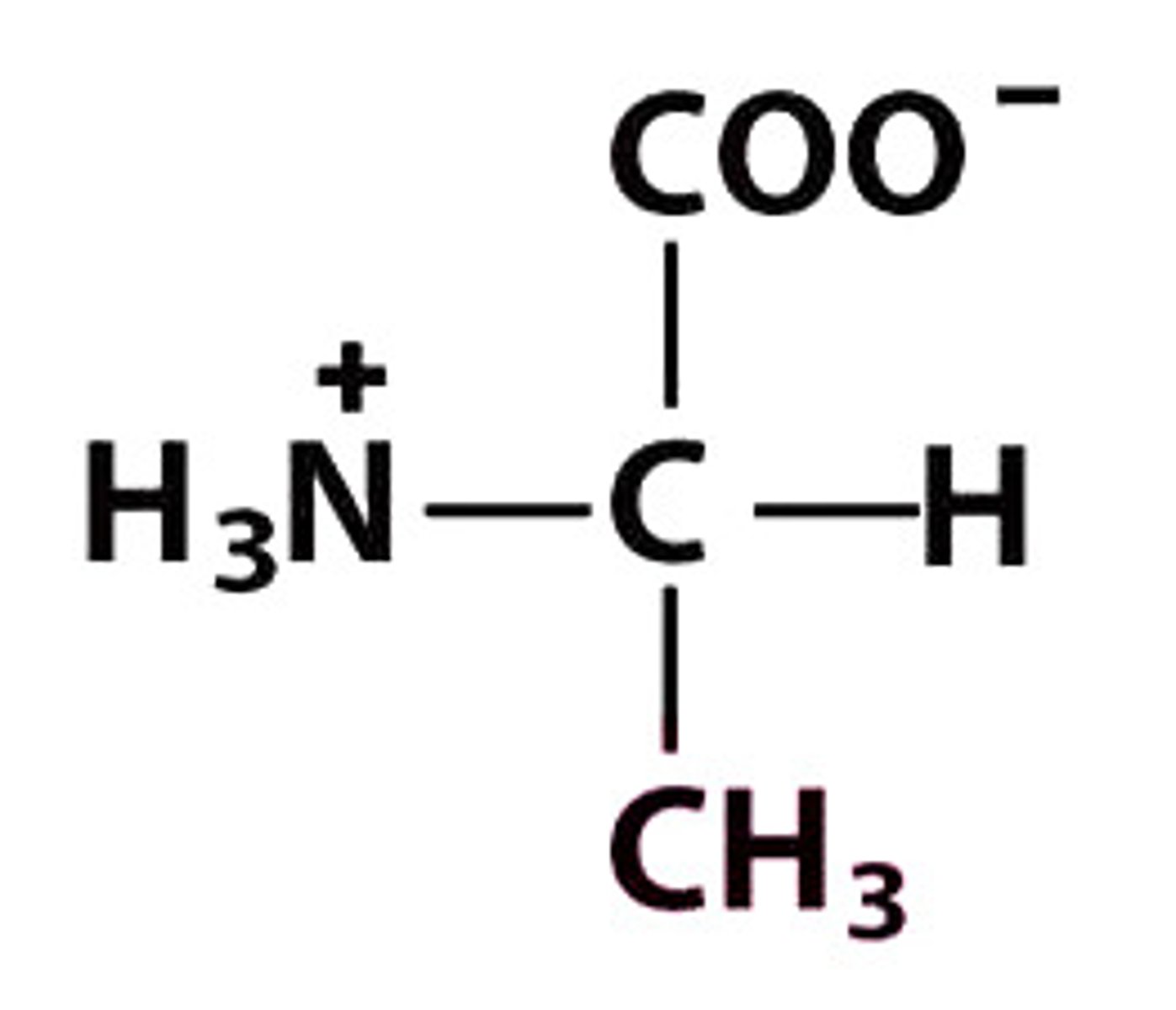
A Ala Alanine
Give the symbol, abbreviation, Name of this amino acid
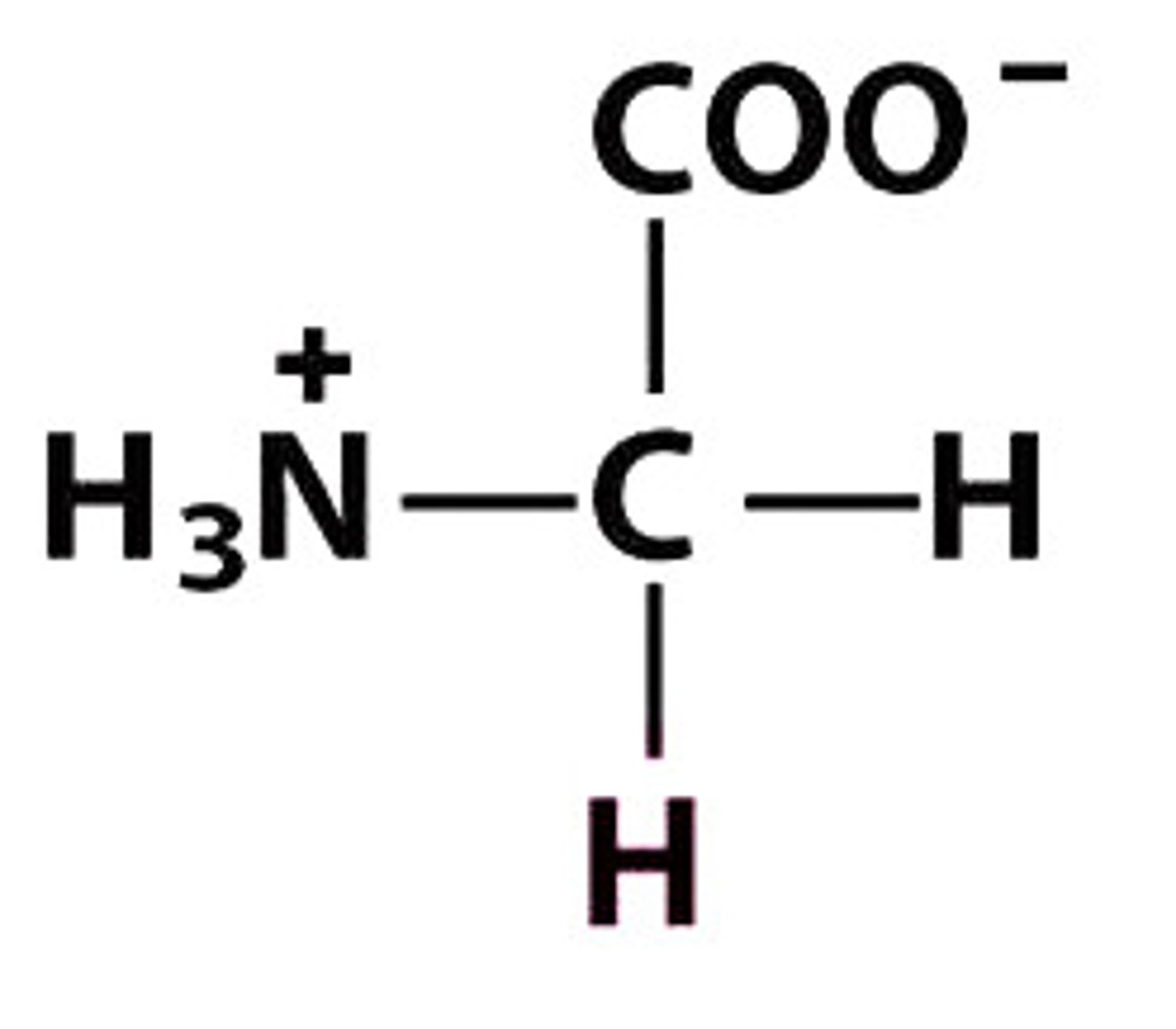
G Gly Glycine
Give the symbol, abbreviation, Name of this amino acid
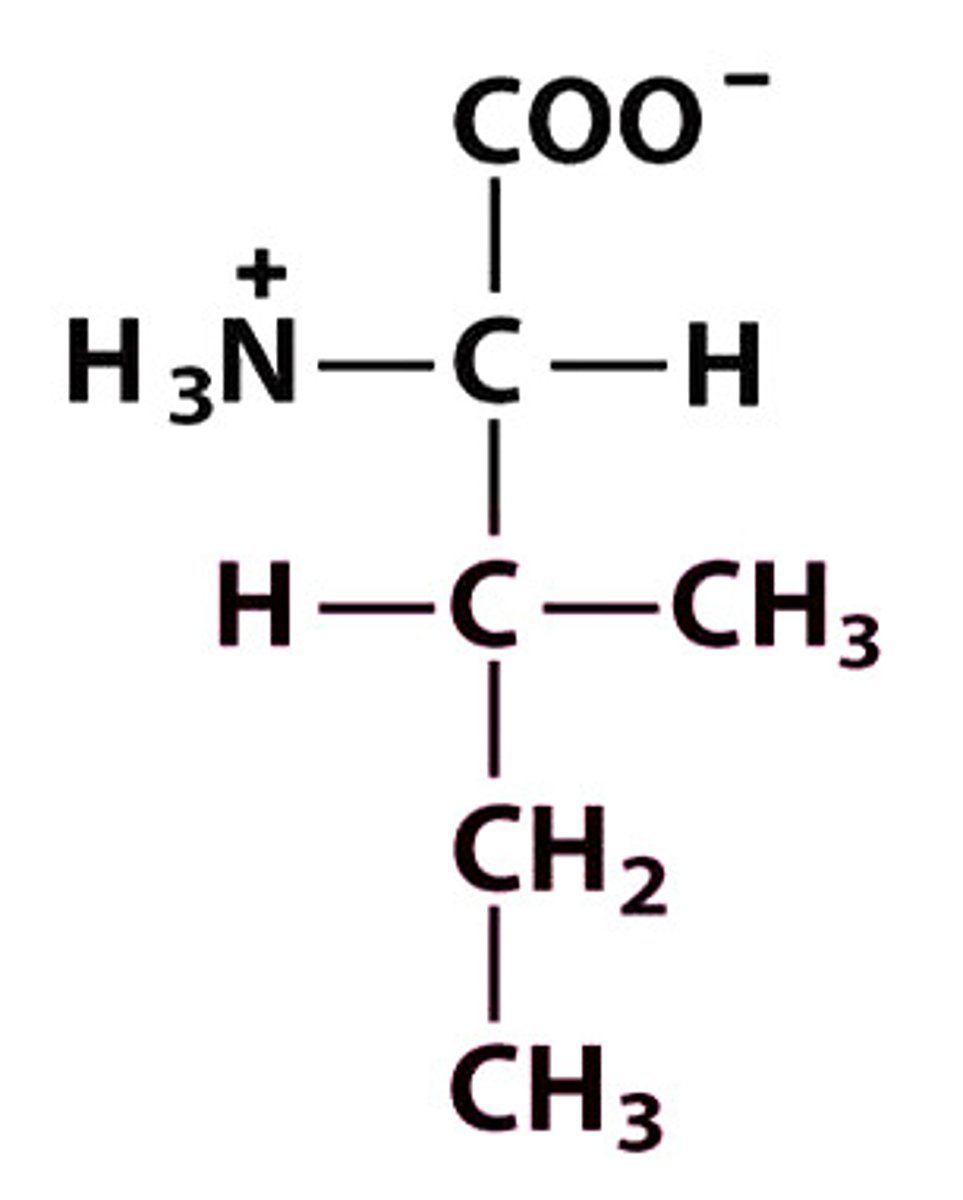
I Ile Isoleucine
Give the symbol, abbreviation, Name of this amino acid
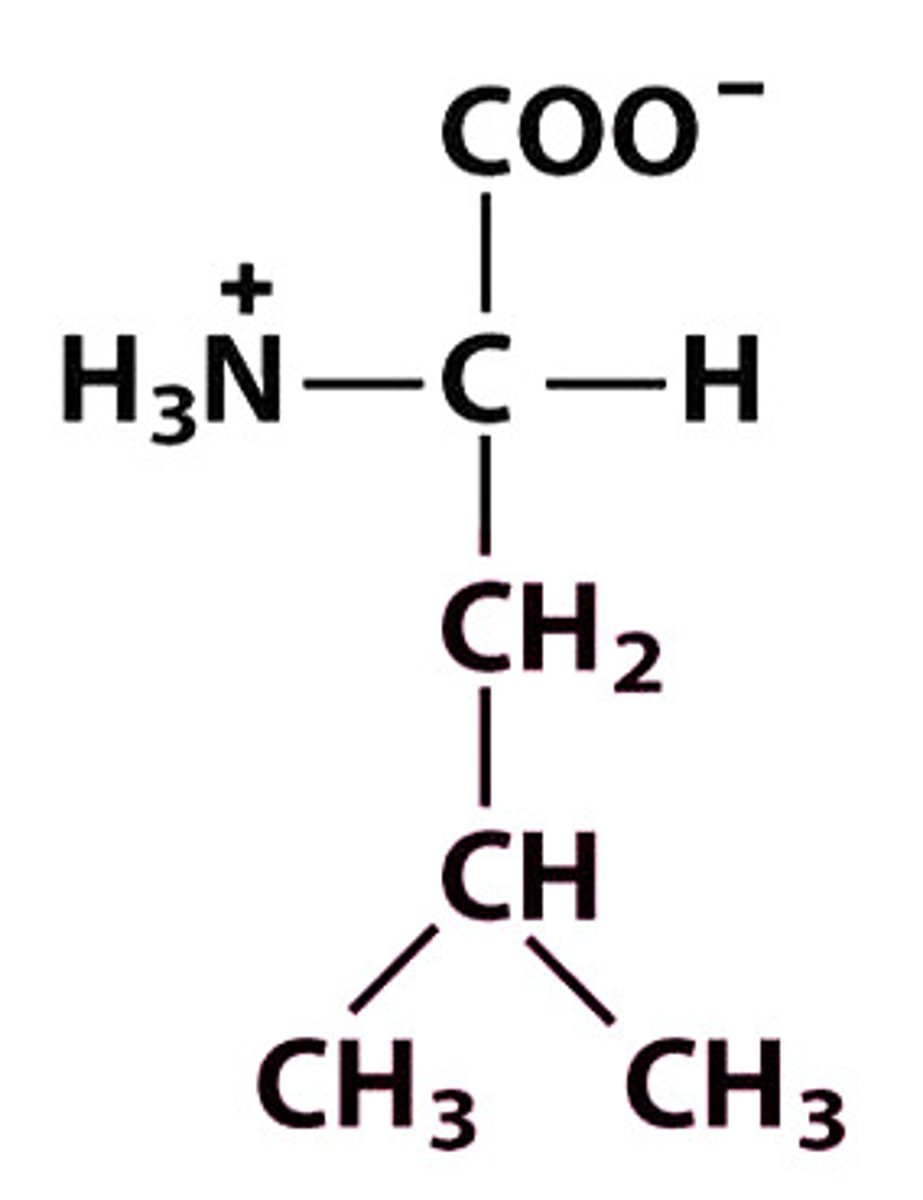
L Leu Leucine
Give the symbol, abbreviation, Name of this amino acid
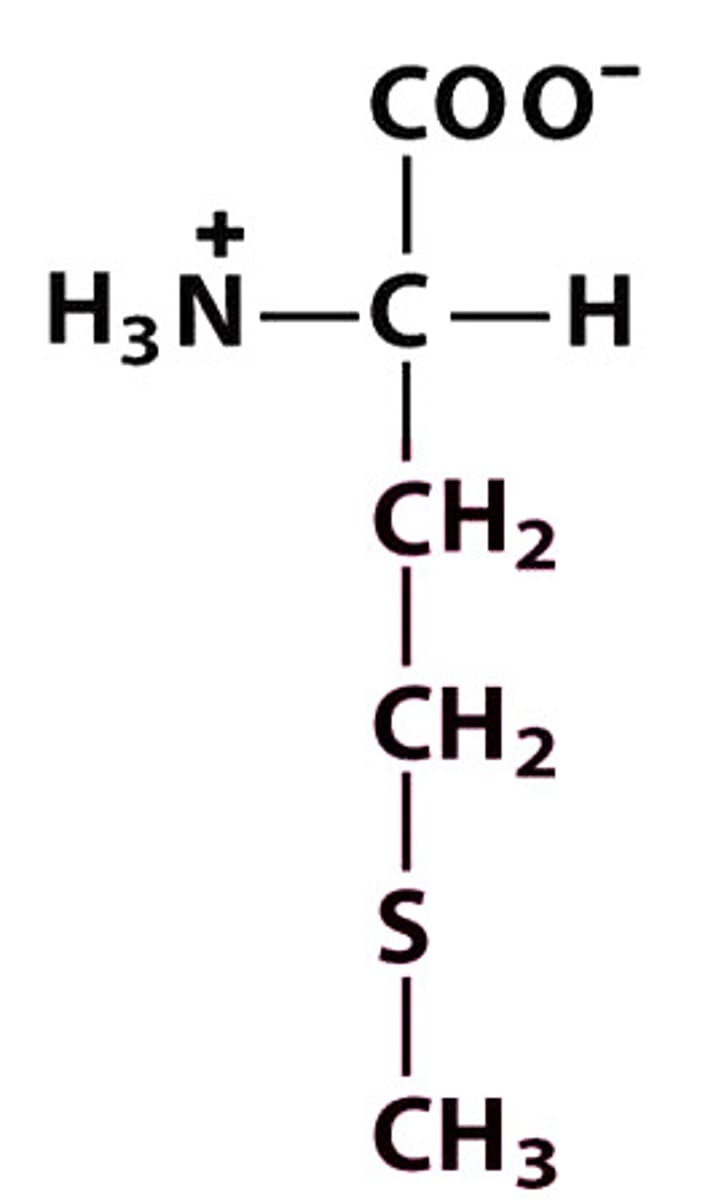
M Met Methionine
Give the symbol, abbreviation, Name of this amino acid
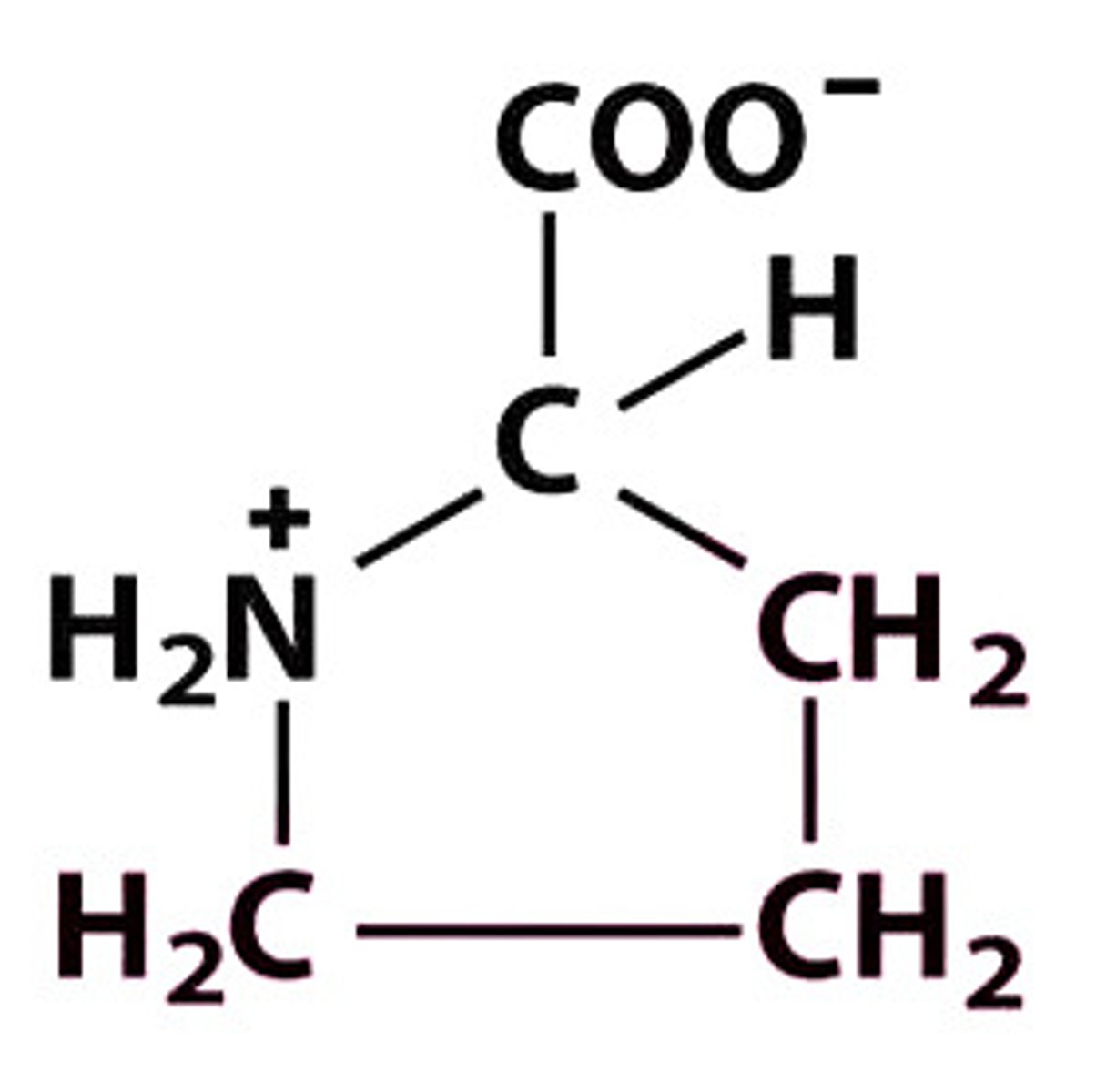
P Pro Proline
Give the symbol, abbreviation, Name of this amino acid
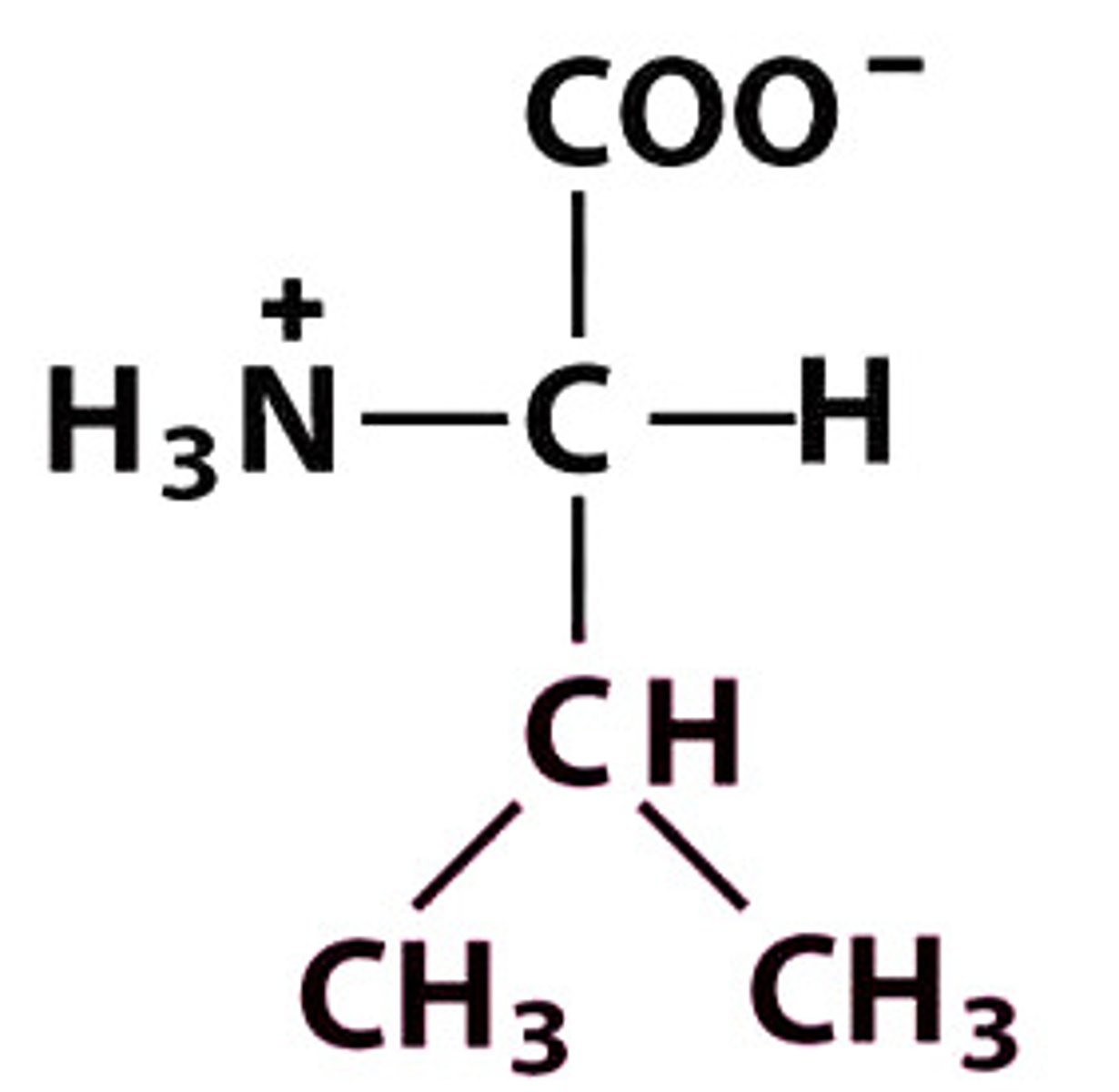
V Val Valine
Give the symbol, abbreviation, Name of this amino acid
Valine, Alanine, Methionine, Proline, Leucine, Isoleucine, Glycine
List the amino acids in the Nonpolar, aliphatic R group
Nonpolar, aliphatic R group
Hydrophobic amino acids often grouped in the middle of a folded protein. Name the Group
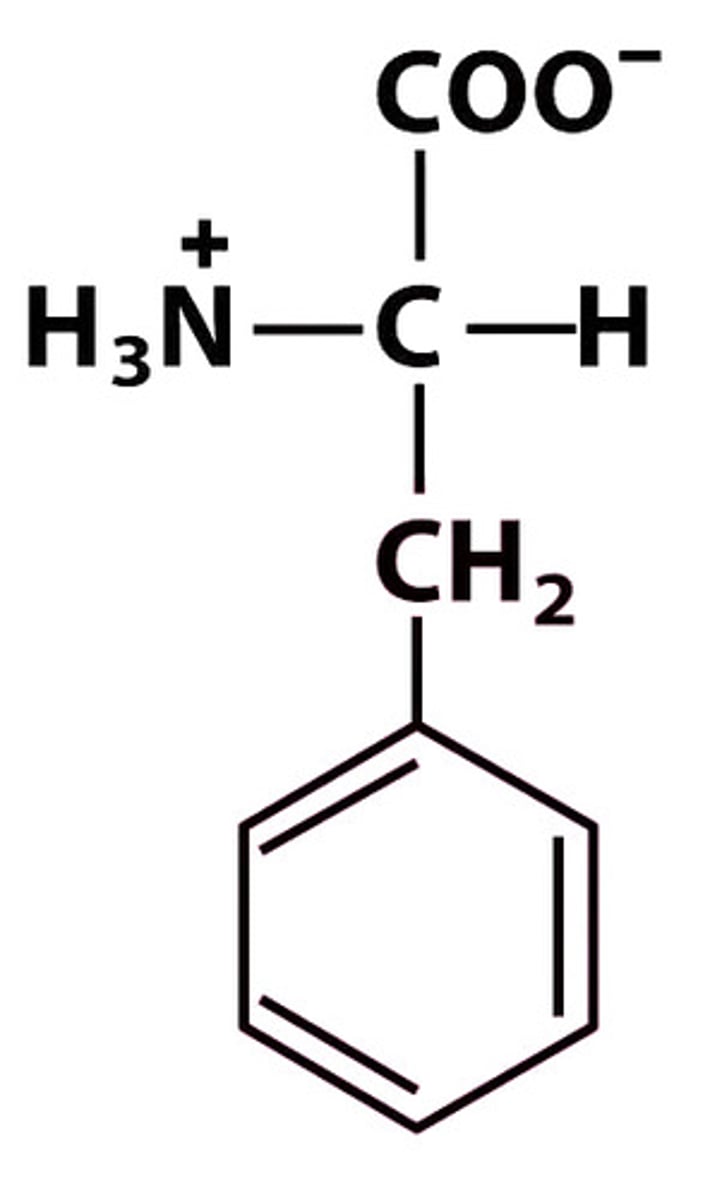
F Phe Phenylalanine
Give the symbol, abbreviation, Name of this amino acid
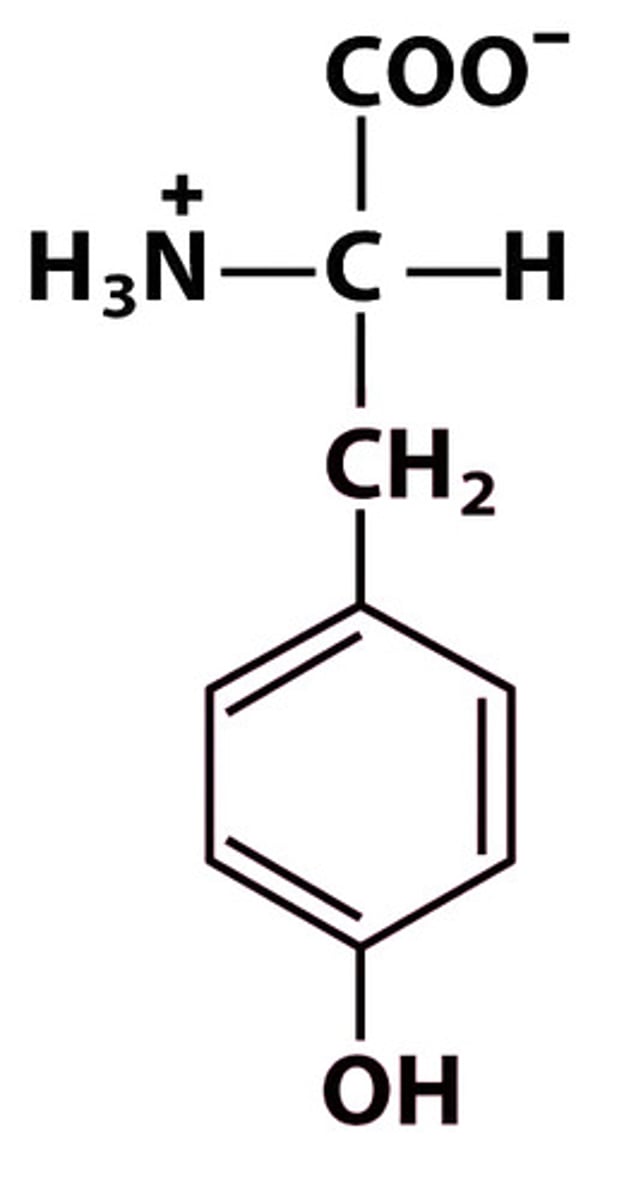
Y Tyr Tyrosine pKa: 10.07
Give the symbol, abbreviation, Name of this amino acid, and identify pka
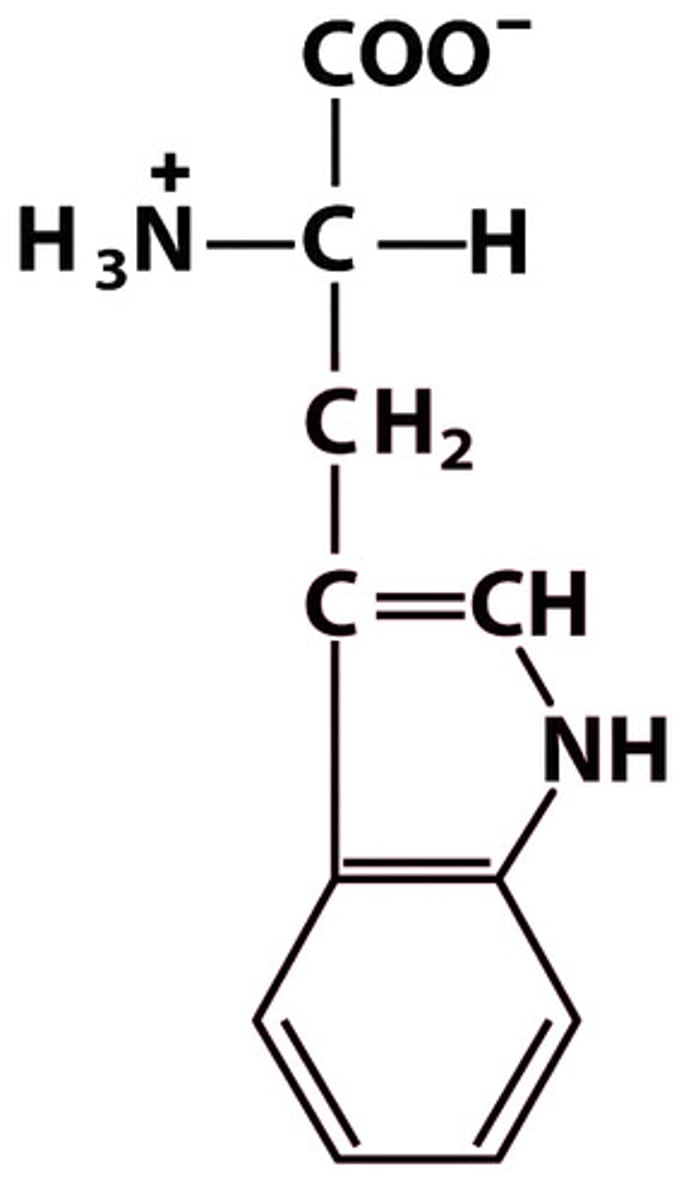
W Trp Tryptophan
Give the symbol, abbreviation, Name of this amino acid
Phenylalanine, Tyrosine, Tryptophan
List the amino acids in the Aromatic R Group
Relatively nonpolar (hydrophobic)
Aromatic R group amino acids are ____
S Ser Serine
Give the symbol, abbreviation, Name of this amino acid
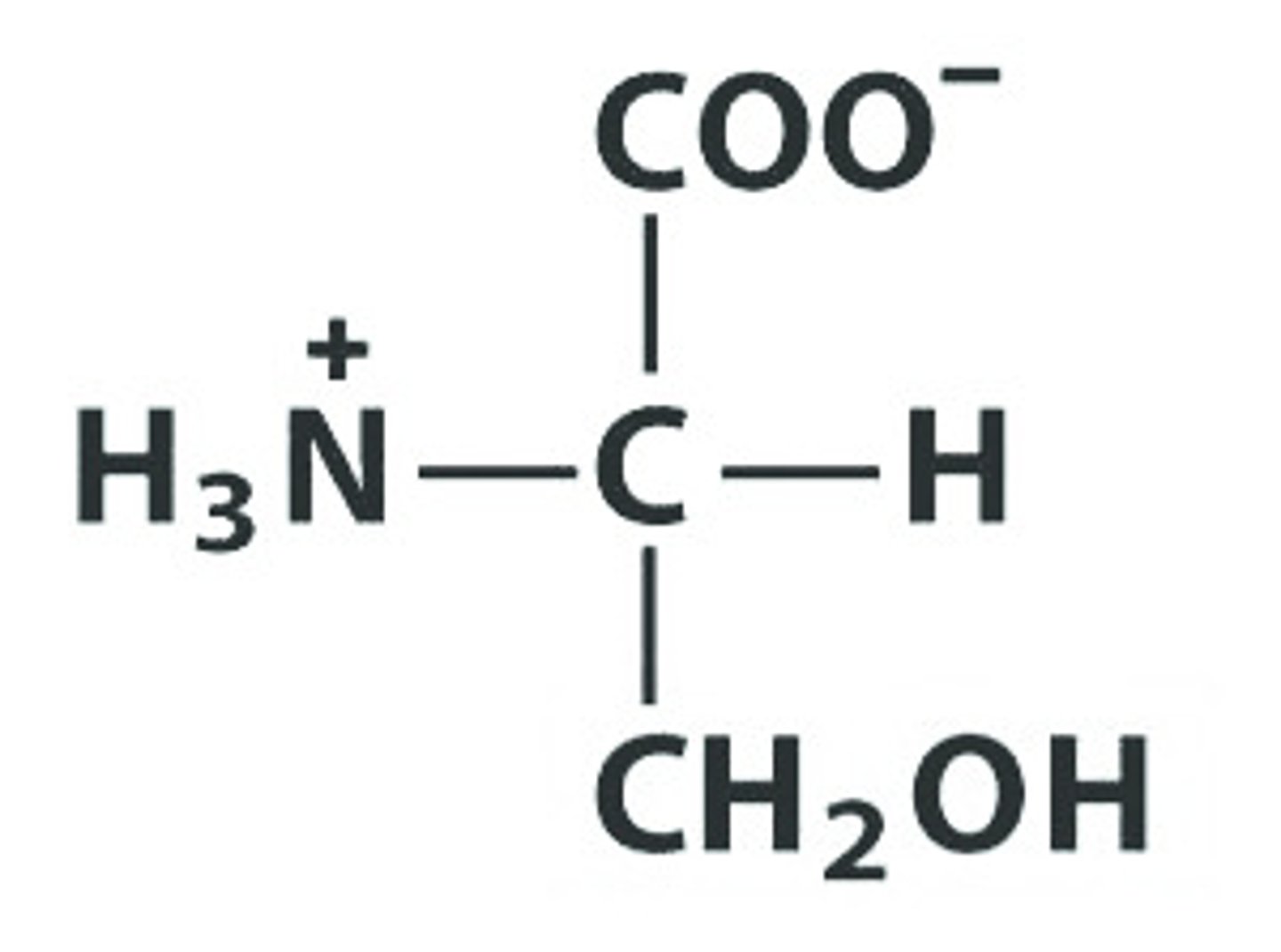
T Thr Threonine
Give the symbol, abbreviation, Name of this amino acid
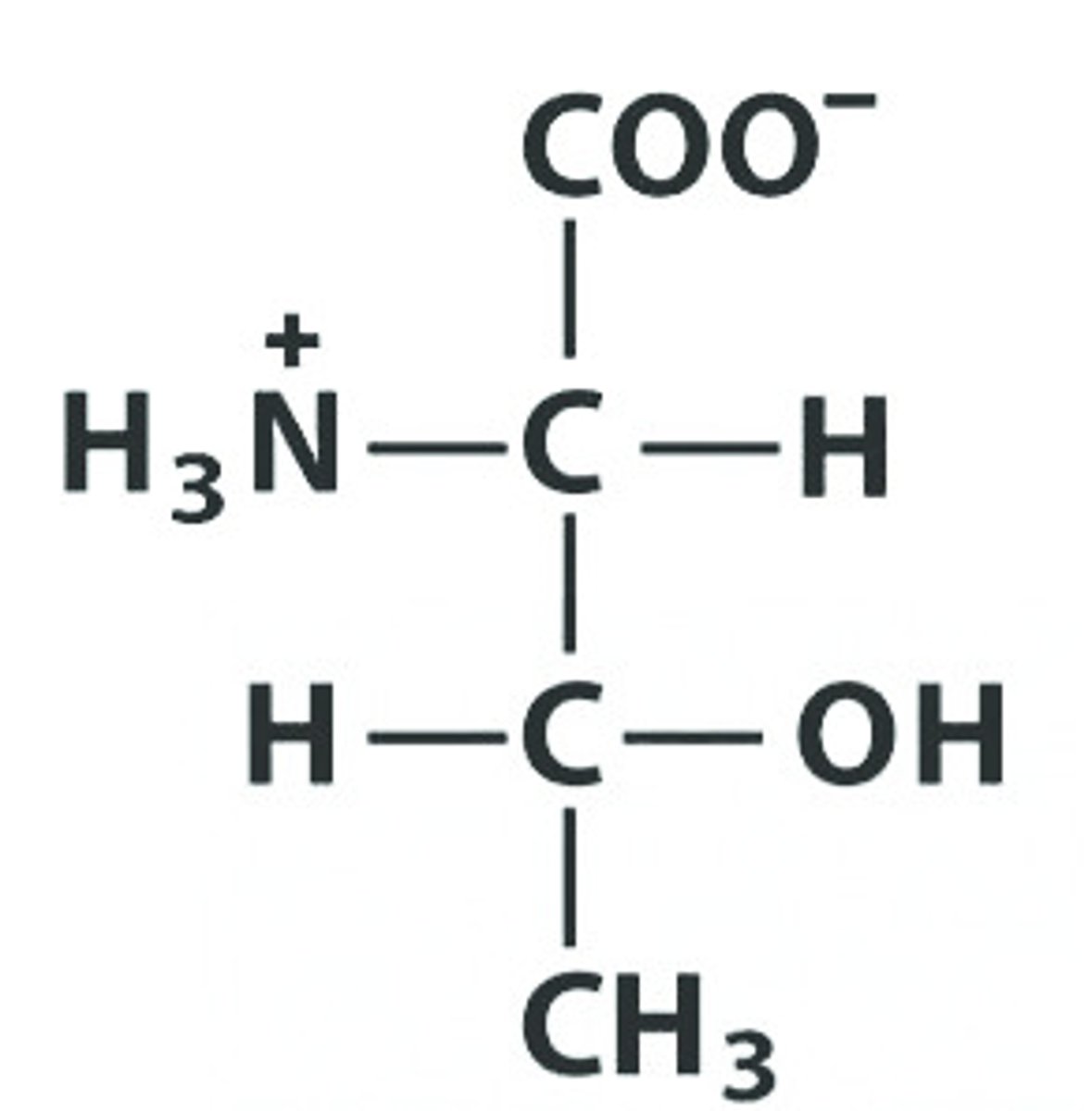
C Cys Cysteine , pKa: 8.18
Give the symbol, abbreviation, Name of this amino acid, identify pka
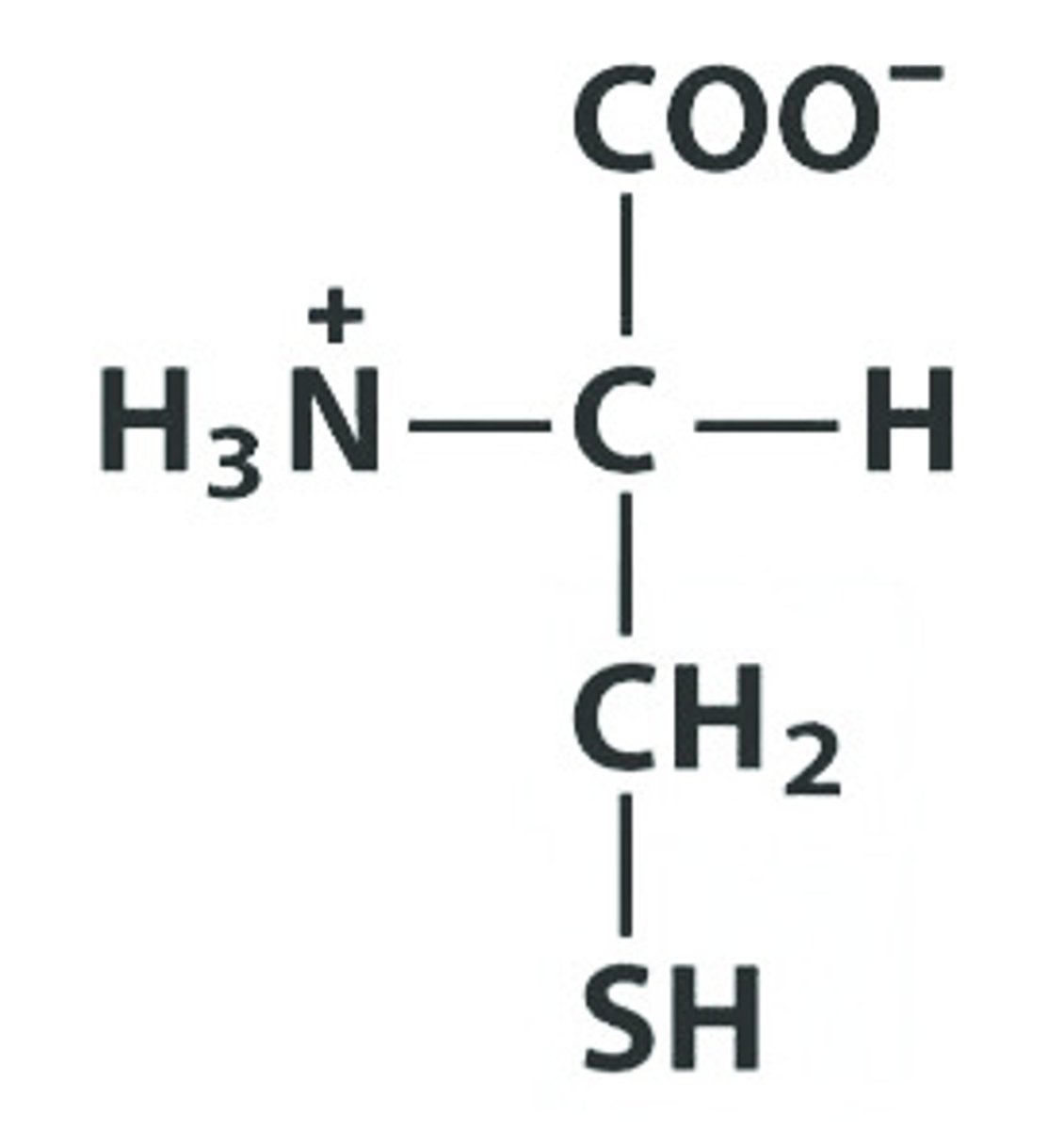
N Asn Asparagine
Give the symbol, abbreviation, Name of this amino acid
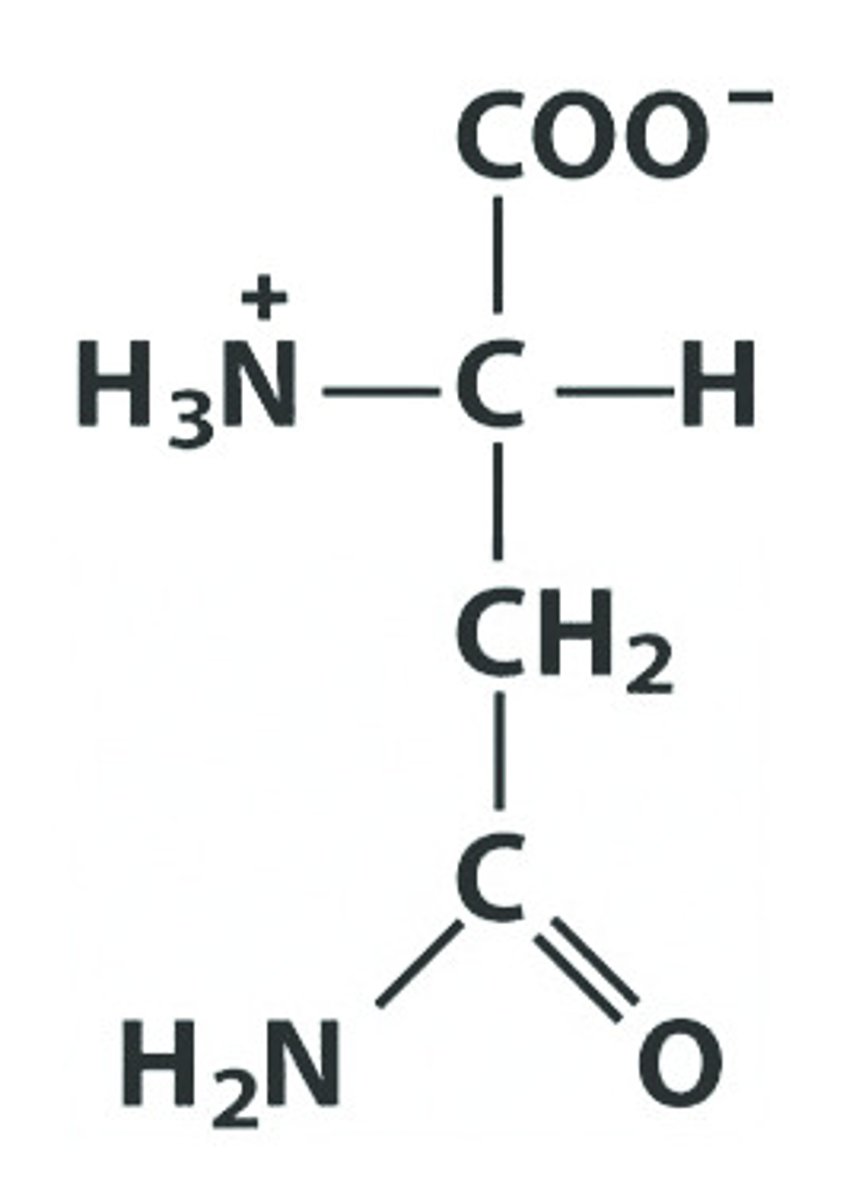
Q Gln Glutamine
Give the symbol, abbreviation, Name of this amino acid
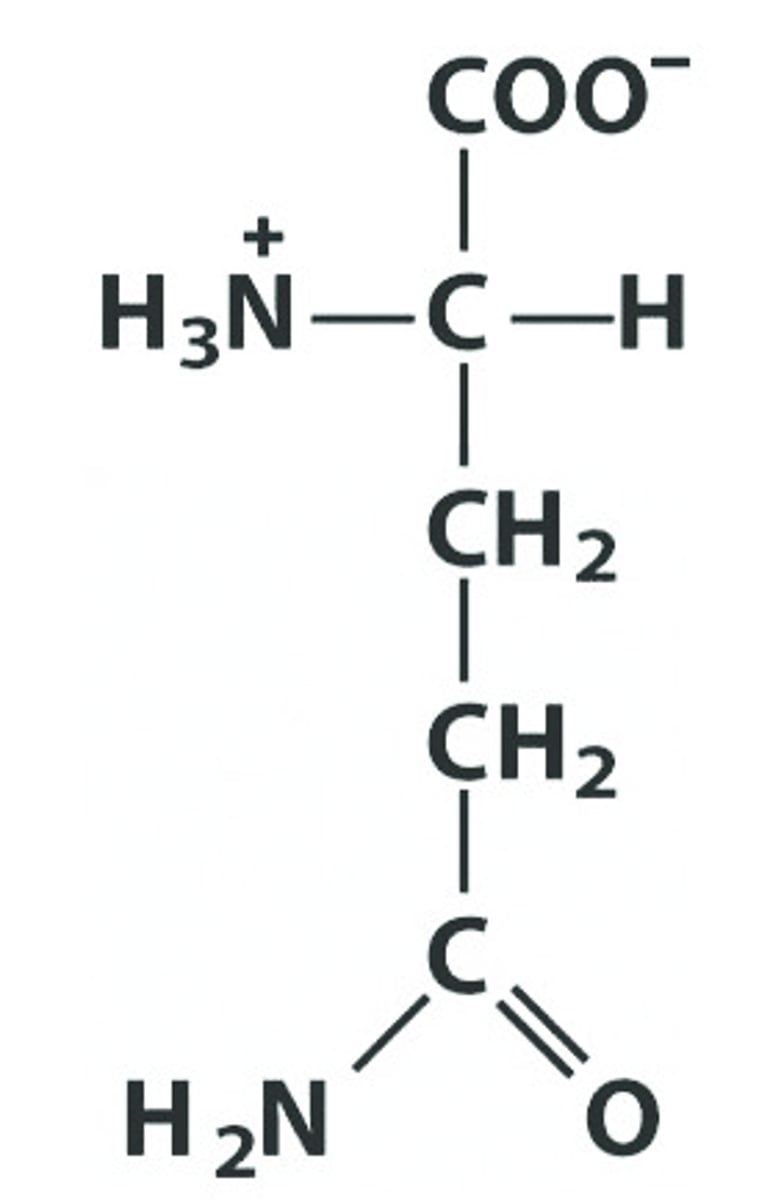
Serine Threonine Cysteine Asparagine Glutamine
List the amino acids in the Polar, Uncharged R Group
K Lys Lysine : 10.53
Give the symbol, abbreviation, Name of this amino acid, identify pka
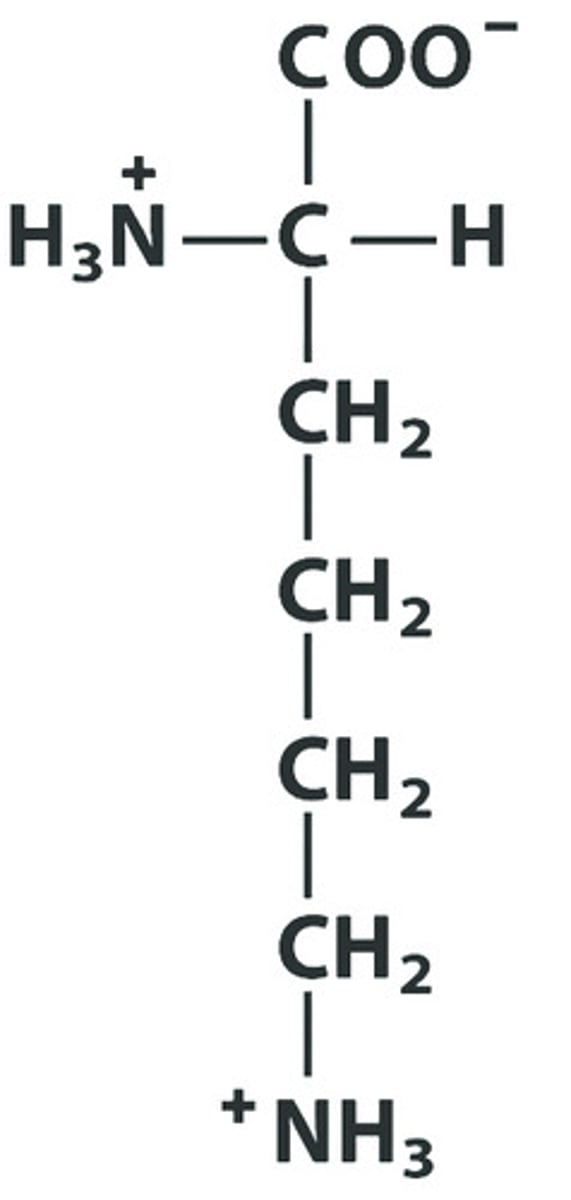
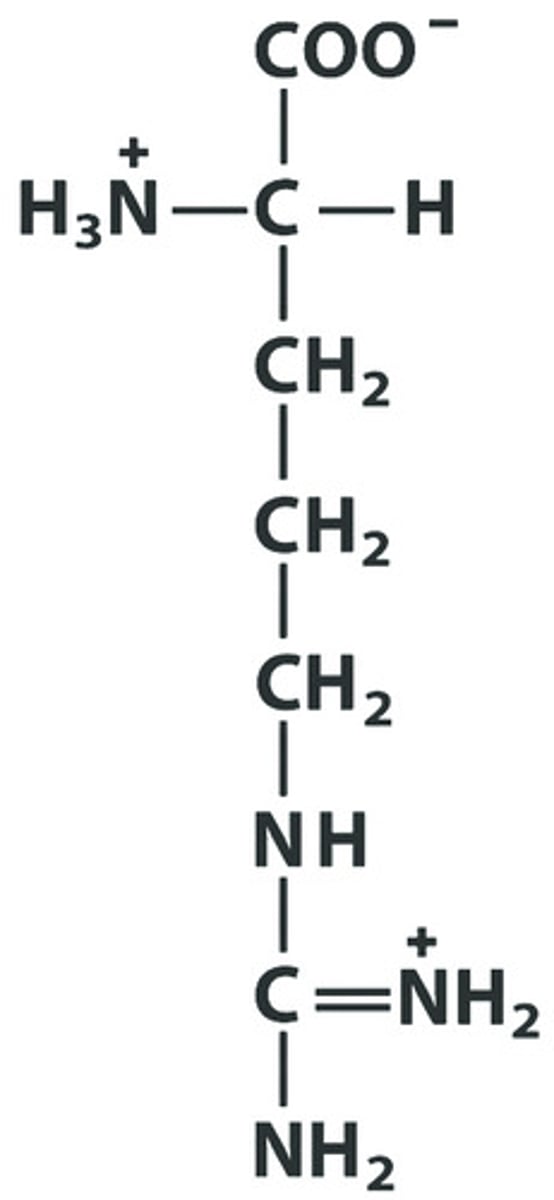
R Arg Arginine pKa: 12.48
Give the symbol, abbreviation, Name of this amino acid, identify pka
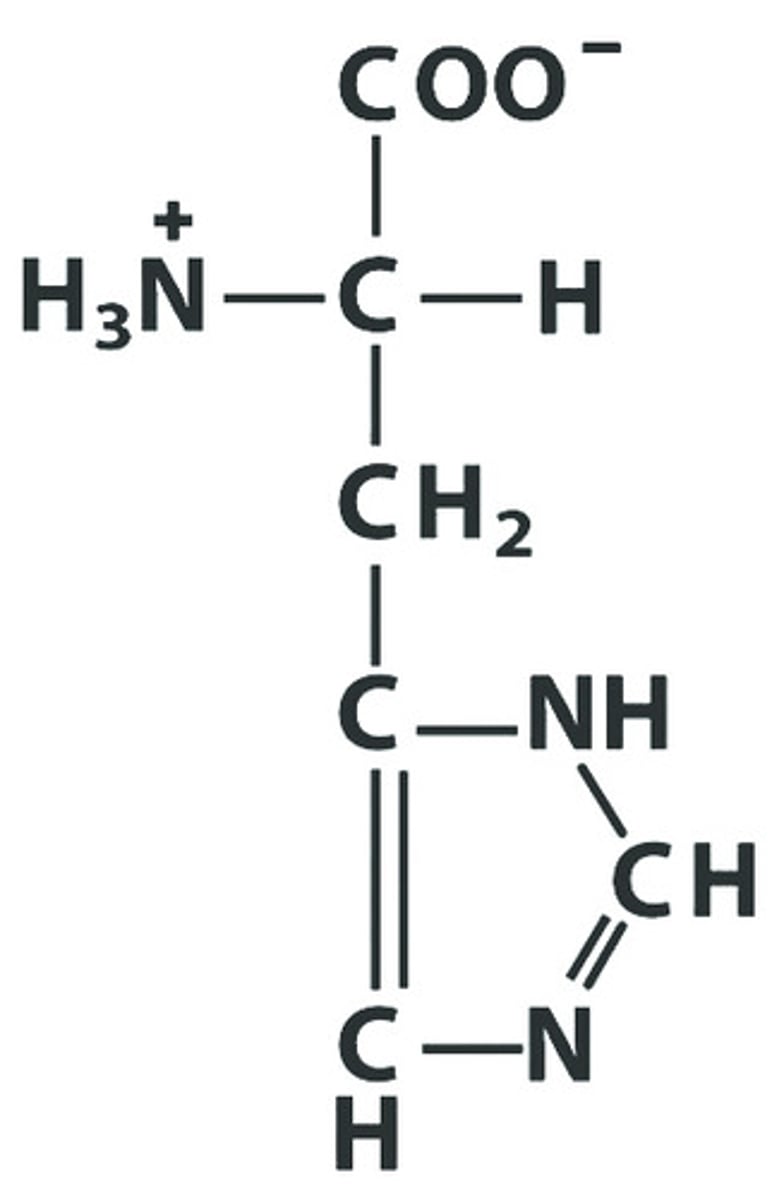
H His Histidine
Give the symbol, abbreviation, Name of this amino acid, identify pka : 6.00
Lysine, Arginine, Histidine
List the amino acids in the Positively Charged R Group
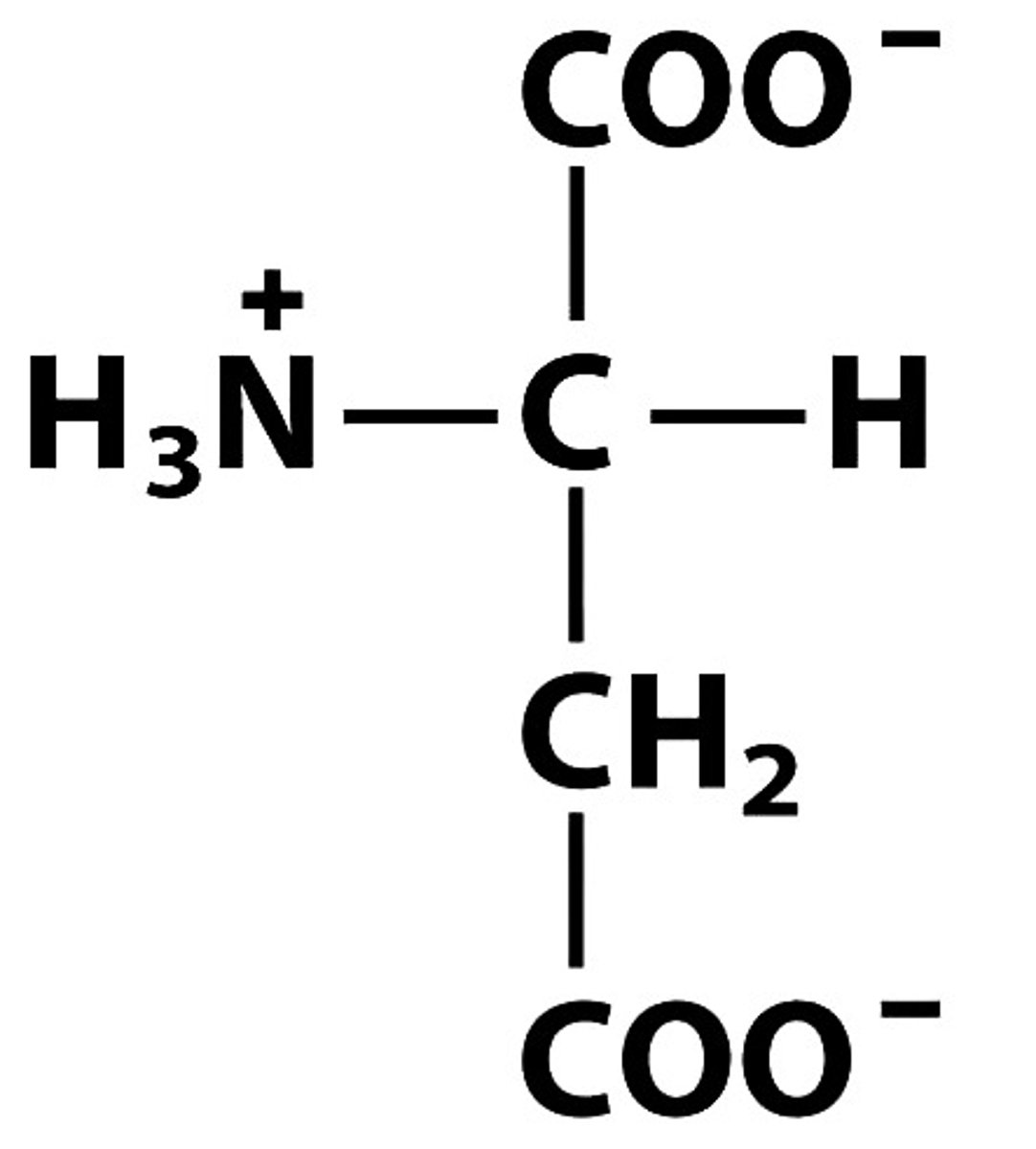
D Asp Aspartate, pKa: 3.65
Give the symbol, abbreviation, Name of this amino acid, identify pka
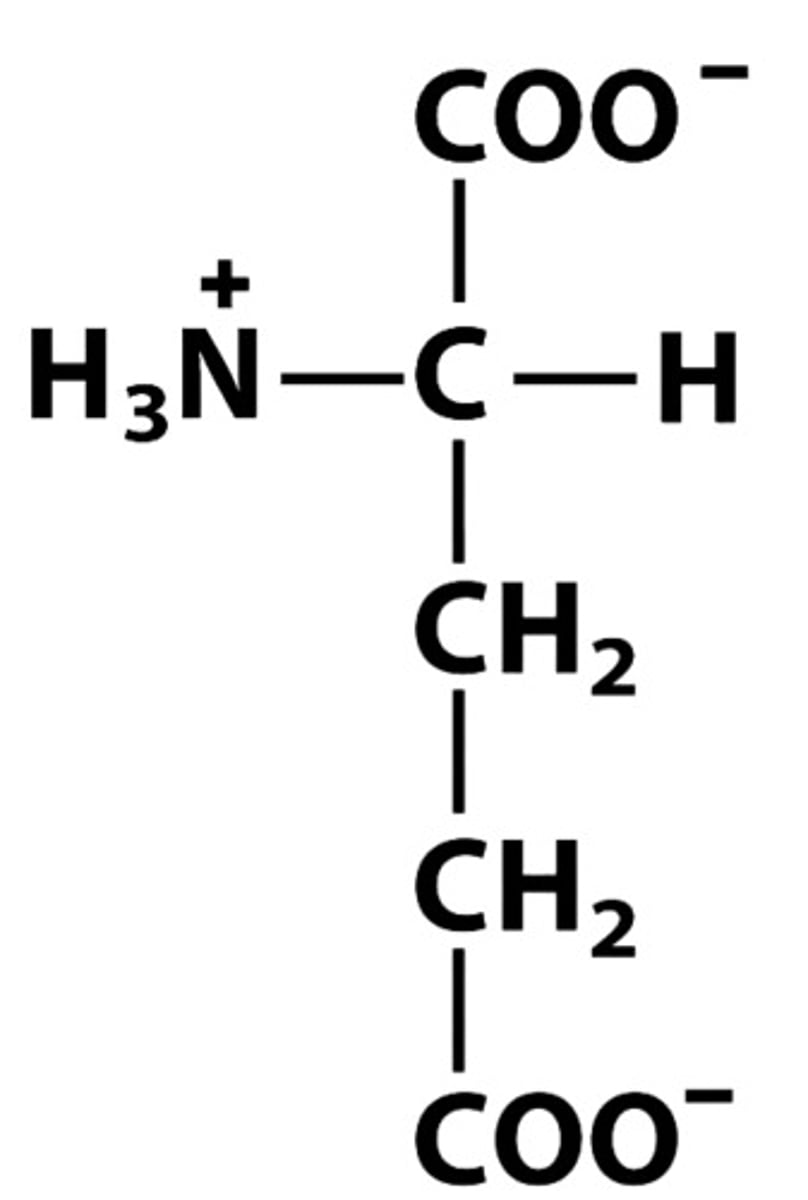
E Glu Glutamate pKa:4.25
Give the symbol, abbreviation, Name of this amino acid, identify pkA
Trypsin
Cleaves at C-side of Lys, Arg (except when followed by Pro)
Chymotrypsin
Cleaves at C-side of aromatic amino acids (Phe, Trp, Tyr).
Pepsin
Cleaves at N-side of aromatic amino acids (Tyr, Trp, Phe) and Leu.
Carboxyl peptidase
Cleaves amino acid at the C-terminus.
Cyanogen bromide
Cleaves at C-terminal side of methionine residues.
Oligomer/Multimer:
Multisubunit protein, with repeating structural units called protomers.
Globular
Folded into spherical/globular shape
Intrinsically disordered proteins
Lack stable tertiary structures, often lack a hydrophobic core, high in charged residues and Pro. Facilitate interaction with multiple partners.
Fibrous proteins
Long strands or sheets, structural function
Edman Degradation
creates phenylthiohydantion PTH dervatives
Dansyl Chloride
Creates Dansyl derivates of the amino aid at the n-terminus
Lithium Borohydride
Followed by acid hydroloysis create alchol derivates at c-terminus. classical used to identify c-terminal amino acids
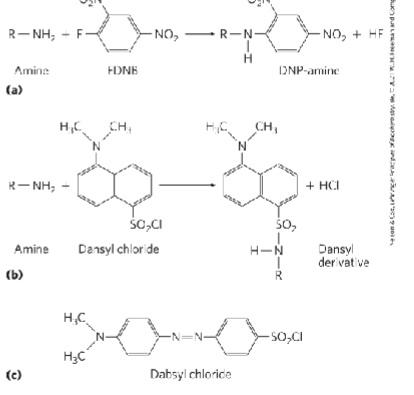
FDNB, dansyl chloride, dabsyl chloride
label amino-terminal a-amino group and e-amino group of lysine residues
Aromatic amino acids
Phenylalanine (Phe, F) → nonpolar, hydrophobic
Structure at pH 7: +H₃N–CH(CH₂–C₆H₅)–COO⁻
Tyrosine (Tyr, Y) → polar (hydrophilic, can H-bond)
Structure: +H₃N–CH(CH₂–C₆H₄–OH)–COO⁻
Tryptophan (Trp, W) → relatively nonpolar but has N in indole ring, amphipathic
Structure: +H₃N–CH(CH₂–indole)–COO⁻
Sulfur-containing amino acids
Cys, C — +H₃N–CH(CH₂–SH)–COO⁻ — polar (forms disulfides)
Met, M — +H₃N–CH(CH₂–CH₂–S–CH₃)–COO⁻ — hydrophobic
Acidic amino acids
Aspartic acid (Asp, D) → acidic, hydrophilic (side chain COO⁻ at pH 7)
Structure: +H₃N–CH(CH₂–COO⁻)–COO⁻
Glutamic acid (Glu, E) → acidic, hydrophilic (side chain COO⁻ at pH 7)
Structure: +H₃N–CH(CH₂–CH₂–COO⁻)–COO⁻
Basic amino acids
Lysine (Lys, K) → basic, hydrophilic (side chain NH₃⁺ at pH 7)
Structure: +H₃N–CH(CH₂)₄–NH₃⁺ –COO⁻
Arginine (Arg, R) → basic, hydrophilic (guanidinium group)
Structure: +H₃N–CH(CH₂)₃–NH–C(=NH₂⁺)–NH₂ –COO⁻
Histidine (His, H) → polar, weakly basic (imidazole ~ partially protonated near pH 7)
Structure: +H₃N–CH(CH₂–imidazole)–COO⁻
Amide amino acids
Asparagine (Asn, N) → polar, hydrophilic
Structure: +H₃N–CH(CH₂–CONH₂)–COO⁻
Glutamine (Gln, Q) → polar, hydrophilic
Structure: +H₃N–CH(CH₂–CH₂–CONH₂)–COO⁻
Branched-chain amino acids (BCAAs)
Valine (Val, V) → nonpolar, hydrophobic
Structure: +H₃N–CH(CH(CH₃)₂)–COO⁻
Leucine (Leu, L) → nonpolar, hydrophobic
Structure: +H₃N–CH(CH₂–CH(CH₃)₂)–COO⁻
Isoleucine (Ile, I) → nonpolar, hydrophobic
Structure: +H₃N–CH(CH(CH₃)–CH₂–CH₃)–COO⁻
Cyclic saturated secondary amino acid
Pro, P — +H₂N⁺ (in pyrrolidine ring) –CH–COO⁻ — hydrophobic
Paper Electrophoresis
Separation of amino acids/peptides on paper based on charge; molecules migrate in an electric field according to net charge at a given pH.
Dialysis
Separates small molecules from macromolecules using a semipermeable membrane; diffusion allows salts/small solutes to pass while proteins/DNA stay inside
Ion-Exchange Chromatography
Elution achieved by increasing salt concentration or changing pH.
Gel Filtration Chromatography (Size-Exclusion)
Separates proteins based on size; larger molecules elute first because they bypass pores, while smaller molecules are delayed.
Affinity Chromatography
Separates proteins by specific binding interactions (e.g., antibody–antigen, substrate–enzyme). Protein of interest binds to a ligand immobilized on the matrix.
Immobilized Metal Affinity Chromatography (IMAC/His-tag purification)
Uses Ni²⁺, Co²⁺, or Zn²⁺ to bind histidine-tagged proteins. Bound proteins are eluted by lowering pH or adding imidazole.
SDS-PAGE (SDS–Polyacrylamide Gel Electrophoresis)
Denatures proteins with SDS, giving uniform negative charge; separates by molecular weight. Smaller proteins move faster through the gel.
Isoelectric Focusing
Separates proteins based on isoelectric point (pI). Proteins migrate in a pH gradient until they reach their pI (net charge = 0).
2D Gel Electrophoresis
Combines IEF (first dimension) and SDS-PAGE (second dimension) to separate proteins by both pI and size.
X-ray Diffraction / Protein Crystallography
Determines 3D structure of crystallized proteins. X-rays diffract off electron clouds; Fourier transform gives electron density map.
NMR Spectroscopy
Studies proteins in solution. Uses nuclear spin (¹H, ¹³C, ¹⁵N, etc.) to give structural and dynamic information, including conformational changes.
Cryo-Electron Microscopy (Cryo-EM)
Visualizes biomolecules frozen in vitreous ice. Useful for large complexes and membrane proteins without need for crystallization.
Electrospray Ionization Mass Spectrometry (ESI-MS)
Produces charged protein/peptide ions in gas phase; measures mass-to-charge ratio (m/z) for molecular weight determination.
MALDI-MS (Matrix-Assisted Laser Desorption Ionization)
Soft ionization technique for peptides/proteins. Sample co-crystallized with matrix, hit with laser, ions measured by time-of-flight MS.
LC-MS/MS Peptide Sequencing
Uses liquid chromatography coupled with tandem MS. First MS separates peptides, second MS fragments them to determine amino acid sequence.
MS/MS Cleavage
Fragmentation pattern in tandem MS used to deduce peptide sequence.