Lecture 3: Recognition of Antigen by B-cells and Antibodies
1/48
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
49 Terms
Basic classification of lymphocytes
B-lymphocytes (B-Cells) → produce antibodies and attack extracellular pathogens - have BCRs
T-lymphocytes (T-Cells) → attack intracellular pathogens - have TCRs
They have receptors on their surface that can recognise foreign molecules, e.g. those on pathogens (antigens)
Origin of B-Cells
Originate and differentiate in the adult bone marrow
Migrate to the lymphoid tissue such as spleen, and mainly the lymph nodes where they become specialised and form plasma B-cells
Origin of T-Cells
Originate in bone marrow
Differentiate to mature T-cells in the thymus before doing their function
Migrate to the lymphoid tissue such as spleen, and mainly the lymph nodes
Adaptive Immune System
It is much more specifc than innate immunity and is driven by
B cells = B cell receptor (BCR)
(membrane-bound antibody).
T cells = T cell receptor (TCR)
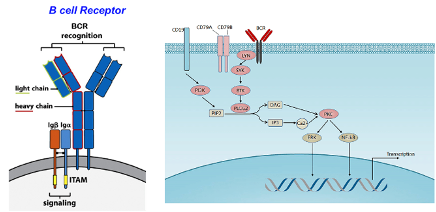
B-Cell Receptor (BCR)
an antibody that is still bound to the membrane – Y-shaped with a membrane domain and adaptor proteins with intracellular domains that can signal the cell
This is important for modulating gene expression, adhesion or survival.
Its signalling determines the fate of antigen-encountered B cells.
Differentiate into plasma B cell
Fate of Antibodies Produced By B-Cell
Antibodies are either:
(i) Attached to B-cell surface as B-cell receptors (BCRs)
(ii) Secreted into plasma (not attached)
BCRs are essential for cell signalling and differentiation
When a BCR binds an antigen:
The B-cell proliferates
Some cells become plasma cells that secrete antibodies
Others become memory B-cells
Main Function of Antibodies
Bind foreign antigens encountered by the host
Mediate effector functions to neutralise or eliminate foreign invaders
Structure of Antibody
Antibody (Ab) = Immunoglobulin (Ig)
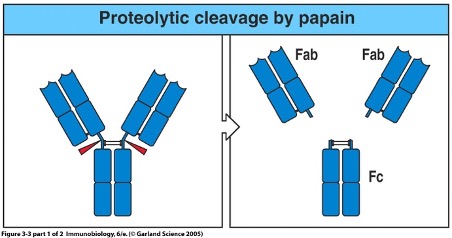
Structure studied using proteases like papain
Papain cleaves at disulphide bonds between heavy chains, producing:
Two Fab (Fragment antigen-binding) regions → bind antigen (N-terminal end)
One Fc (Fragment crystallisable) region → does not bind antigen, involved in immune functions (C-terminal end)
Antibody Structure: Heterodimers
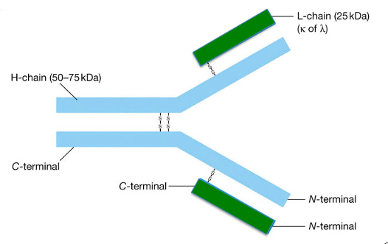
They have:
two identical light chains (L-chain) - smaller
two identical heavy chains (H-chain) – bigger
Chains are bound together by disulfide bonds
Antibody Structure: Immunoglobulins
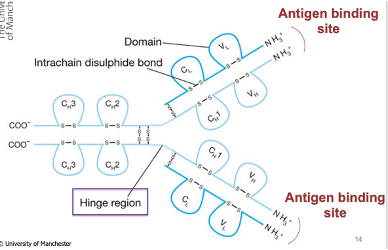
They have immunoglobulin domains that give rise to the final structure of the Ab
They have a hinge region that allows flexibility in movement to allow binding to antigens
The antigen binding site of Abs is located at the N-terminus
Antigen
A (general) molecule recognised by an antibody – not PAMPs (they are shared by many pathogens)
Usually big molecules (>4000Da)
Proteins (most common)
Carbohydrates (Polysaccharides)
Lipids
DNA
Antigenic Epitope
The parts of the antigen recognised by antibodies -small fragments that are recognised by the Ab
e.g. a virus is an antigen; the Ab recognises a small part of the virus
Each antigen has hundreds → between 1011 - 1025
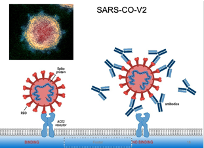
Antigenic Epitope of SARS-COV2
Antibodies generated by infection/ vaccine bind to the spike protein of Sars and not the whole virus
Antigen-Antibody Interaction
Antibody interacts with a specific binding site which is determined by the hypervariable regions of the antibody
antigen binding site must be complementary in shape to the antigenic epitope
Hypervariable regions form the complementary determining region (CDR), allowing for specific recognition
CDR regions undergo gene rearrangement, enabling the production of diverse antibodies to recognise many epitopes
Binding requires shape complementarity between the CDR (Ab) and epitope – they must fit well together to have an effect
it is specific
Variable vs Constat Regions of Antibodies
Variable regions are located at the N-terminal end (last domain)
They vary between antibodies and determine antigen specificity
Constant (C) domains remain unchanged across antibodies
Provide structural support and effector function
Each antibody has a unique antigen-binding site shape due to its variable region
Conformational Epitopes
Antigenic epitopes with a 3D shape
Structural, formed by protein folding but may not be a continuous sequence of amino acids.
Most antibodies recognise these
Linear Epitopes
Recognise amino acids in sequence (2D) → Continuous sequence of amino acids.
Rare in nature; seen when proteins are denatured (e.g. Western blots)
Less common.
Antibody Recognition of Antigens & Epitopes
Different antigens/pathogens can share similar surface structures (epitopes), allowing the same antibody to bind to multiple antigens.
An antibody may recognise antigens 1, 2, and 3 if they all share a common epitope.
Some antibodies are specific to unique epitopes on a single antigen.
During infection, the immune system produces many different antibodies, each targeting different epitopes on the antigen.
Antigen-Antibody Interactions: Affinity and Avidity
How strongly an antibody binds to an antigen – want it to bind as strongly as possible so that it is bound for as long as possible to induce the effector function
Non-Covalent Forces
Weak forces that are involved in antigen-antibody binding
Hydrogen bonding
Electrostatic interaction
Van der Waal forces
Hydrophobic interactions
The antigenic epitope must be very close to the Ab to bind and maximise these interactions
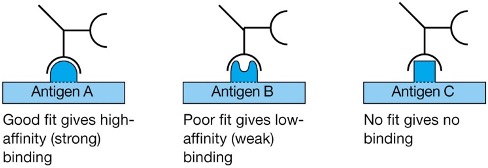
Antigen-Antibody Interactions: Specificity
Different antibodies bind different antigenic epitopes
Affinity
The strength of the binding between an epitope and an antibody-antigen binding site.
Interactions are weak unless the two binding molecules are close together
i.e. need a good fit between antigenic epitope & antibody binding site
Cross-Immunity
When an antibody to one antigen provides protection against another.
This can happen in two ways:
The antibody binds specifically to the same epitope found on different antigens.
The antibody has weaker affinity but still binds sufficiently to similar epitopes.
Even low-affinity binding to an epitope is mportant for cross-immunity.
Cross-Reactivity
The same antibodies could bind different antigenic epitopes
It is good as it can allow one antibody to recognise a variety of pathogens
Affinity for the cross-reacting epitope is less than for the original epitope
It is problematic if an Ab recognises self-proteins via cross-reactivity and low affinity, which may result in an autoimmune response
Avidity
The overall strength of an antibody-antigen complex
If an antibody uses both binding sites to bind to 2 epitopes on the same particle this increases the total binding strength
Affinity vs Avidity
Affinity refers to the strength of binding between a single antibody binding site and an epitope.
Avidity is the overall strength of binding when multiple binding sites are involved.
High affinity, low avidity – antibody binds strongly to one epitope, but only one site is occupied.
High affinity, high avidity – antibody binds strongly and both binding sites are engaged.
How do antibody levels in the body compare to the amount needed for protection?
Number of antibody specificities = 1011
Volume of blood = 5000 ml (5L)
Protective antibody level = 10 ng/ml
Amount of antibody required:
Calculation: 1011 x 5000 ml x 10 ng = 5 x 1015 ng = 5000 kg
Comparison: This is far more than the 70 kg body weight!
The body doesn't produce enough antibodies for constant protection. Antibodies are mainly generated after infection or vaccination, reaching levels that provide protection.
Antibody Production After an Infection
First infection – initial immune response, innate immunity; after day 7 adaptive immunity; after 2 weeks recovery
Following re-infection protective immunity present – little to no symptoms – peak of Ab production
Overtime immunological memory is generated – provides long-term protection – sharper and higher peak of Ab production
Types of Antidbody Response
Acquired – Abs produced after exposure to the pathogen
Specific - for a specific pathogen – go through a process
Memory - 2nd response bigger and faster – longer protection
Key Features of Antibody Structure
Constant domain: Provides structural stability.
Variable domain: Responsible for antigen specificity.
Antigen binding site: Located at the N-terminus of the antibody.
Fc (crystallizable fragment): Allows the antibody to interact with pathogens or antigens. Known as the effector domain.
Humoral Immune Respnse
Mediated by antibodies in serum
Antibodies have many effector functions
Neutralisation
Opsoninatio
Activation of Complement
Antibody dependent cell-mediated cytotoxicity (ADCC)
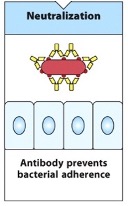
Neutralisation
Antibodies prevent pathogens or proteins from binding to their target.
Mechanism: Antibodies bind to epitopes on pathogens, preventing them from adhering to surfaces or entering cells.
Protection: This is crucial for preventing viral infections, as it stops the virus from entering cells and replicating.
Vaccines: Many vaccines, such as COVID-19 vaccines, induce this response by preventing spike proteins from binding to cell receptors, thereby preventing infection.
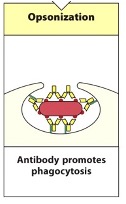
Opsinosiation
A pathogen tagged by antibodies is phagocytosed by phagocytic cells such as macrophages or neutrophils.
Pathogen taken up by phagocytic cell and is placed in a phagolysosome where the pH is low and full of proteases that will destroy the pathogen
The fc-domain of the antibody binds the Fc-receptor on the phagocyte to trigger phagocytosis – triggers the uptake of the pathogen into the phagocytic vesicle
Complement Activation
Antibodies attached to the surface of a pathogen activate the first protein in the complement system.
This can lead to two things:
Complement proteins (C5b) deposit on the pathogen's surface, leading to pore formation, altering osmolality, with fluid entering the pathogen, and causing the pathogen to lyse (burst).
mediated by Membrane attack complex
Complement receptors on phagocytes recognise the deposited complement proteins, inducing phagocytosis. The antibody's Fc region binds to receptors on the phagocyte, triggering receptor oligomerization, which facilitates pathogen uptake into phagolysosomes for destruction.
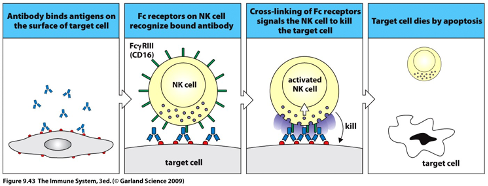
Antibody-Dependent Cell-Mediated Cytotoxicity (ADCC)
Host cells infected by viruses can express viral proteins on their surface, which helps the virus evade detection.
Antibodies specific to the virus recognise and bind to these viral proteins.
NK cells (innate immune cells) and macrophages have Fc receptors (FcR) that can bind to the antibody bound to the infected cell.
Upon binding, NK cells release perforin (a lytic enzyme) and granzymes that mediate extracellular killing and induce cell death by apoptosis, leading to the removal of infected cells without phagocytosis.
This process is key for eliminating intracellular pathogens, especially viruses, by indirectly targeting infected cells for destruction.
Immunotherapies
Can generate antibodies against certain abnormal proteins expressed by cancer cells, triggering the toxicity-dependent effects to kill specific target cells
This can be done by directly triggering the tumour cell to be coated with the Ab, ADCC/NK cell response, with it binding to the receptor of the Ab generated to target these cells, which may induce phagocytosis
Agglutination- Blood Typing:
Ability of an antibody to immunoprecipitate a soluble or non-soluble antigen.
Agglutination occurs where blood cells precipitate and break – used to identify blood types
A and B transferases are encoded by the ABO gene
Blood groups: A, B, O
Group A express the A transferase antigen; Anti-B Abs in serum – if receive type B blood – attack B-transferase as if it was a threat
Blood Group Genotypes:
OO: O
AA, AO: A
BB, BO: B
AB: AB
Antibody Classes
They are determined by different C regions in the heavy chain of the antibody.
There are 5 types
Each has unique structural and functional characteristics that allow them to perform specific functions.
Functions:
Bind to Fc receptors.
Activate complement.
Regulate secretion.
An antibody can be of a different class but still have the same antigen-binding specificity.
Different antibody classes are identified based on their Fc receptor.
IgG Antibody
The most abundant
4 classes
Ig_1 IS THE MOST COMMON
IgM Antibody
Earliest antibody made after antigen contact
Pentamer via J chain and disulphide bonds
Up to 10 binding sites
IgD Antibody
Rare
function in serum is unclear
IgA1 Antibody
Serum and major Ig in Mucosa
2 subclasses
Dimeric structures in secretions → 2 Ig molecules joined together by the J chain
IgE Antibody
Lowest present in serum
Bind to mast cells
Important in Allergy and worm infection
IgM Polymer
Forms a pentamer with 5 Ig_’s binding through disulfide bonds and a J-chain (15kDa peptide chain)
It is able to bind an epitope more strongly due to the formation of a complex that is able to bind more domains
IgA Polymer
Forms a dimeric structure with the assistance of the J-chain, which is key for its function
Immunoglobulin A
Produced by B cells in the bone marrow and then moves to secondary lymphoid tissues.
Another major site of production is Mucosal Associated Lymphoid Tissue (MALT), such as in the gut.
~approximately 5g/day produced
Predominantly found in external secretions like saliva, tears, and mucus.
It serves an important role at membrane surfaces, which are the main entry points for pathogens.
Helps prevent pathogen entry by acting as a barrier.
a dimer in secretions, joined by a J chain.
IgA Entry Into The Gut
IgA antibodies form a dimer.
A molecule called the poly Ig receptor is found on the basolateral side of intestinal epithelial cells.
This receptor attaches to IgA, and is internalized into the cell.
It is then transported to the lumen of the gut.
Secretory Component
Part of the poly Ig receptor is cleaved, and the remaining portion becomes the secretory component (SC).
SC protects IgA from being destroyed by gut enzymes.
SC allows IgA to be transported into the gut.