Exam Four
1/320
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
321 Terms
How do cells determine their function if every cell except gametes and red blood cells have the same DNA?
Gene expression determines cellular function.
How is gene expression regulated in prokaryotes?
Since prokaryotes lack a nucleus, gene expression is primarily controlled at the level of transcription through mechanisms such as operons and transcription factors.
What is the first layer of regulation in a eukaryotic organism for gene expression?
Chromatin because the DNA has to be accessed
What is the second layer of regulation in a eukaryotic organism for gene expression?
Transcription
What is the third layer of regulation in a eukaryotic organism for gene expression?
RNA processing
What is the fourth layer of regulation in a eukaryotic organism for gene expression?
Translation because of the interaction of RNA with the Ribosome.
What is the fifth layer of regulation in a eukaryotic organism for gene expression?
Post-Translation because of alterations in protein product.
There are two major protein classes to regulate chromatin decondenstion: ATP-Dependent Chromatin Remodeling Complexes, and Histone Modifying Complexes. What is ATP-Dependent Remodeling Complexes?
These are multiprotein complexes that utilize ATP hydrolysis to reposition or restructure nucleosomes, allowing for increased accessibility of DNA for transcription and other processes. Done through the sliding or eviction of nucleosomes.
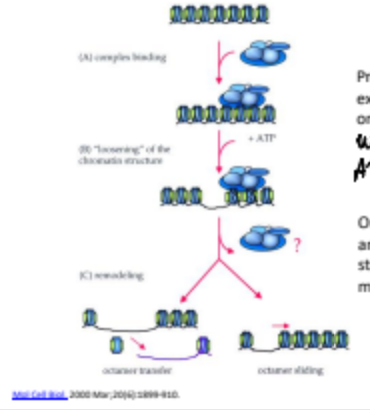
There are two major protein classes to regulate chromatin decondenstion: ATP-Dependent Chromatin Remodeling Complexes, and Histone Modifying Complexes. What are histone/DNA modifying complexes?
These complexes chemically modify histones or DNA, influencing chromatin structure and altering gene expression by adding or removing functional groups such as acetyl or methyl groups.
If Acetylation occurs what happens to the DNA?
It is expressed
If Methylation occurs, what happens to the DNA?
It is usually repressed.
What protein performs methylation?
Methyltransferases, which add methyl groups to DNA or histones, typically silencing gene expression.
What is the Histone Code Hypothesis?
The hypothesis that transcription of DNA is regulated in part by specific chemical modifications to histone proteins. Modification to the histone proteins themselves recruit other proteins to the modified histone. The recruited proteins act to alter chromatin structure or affect transcription.
What is the first step of signaling control of gene expression in the transcription step?
The presence of an initiating signal
What is the Second step of signaling control of gene expression in the transcription step?
Signal pathway cascade. A series of molecular events triggered by the initiating signal that ultimately lead to changes in gene expression.
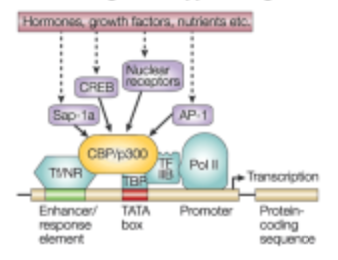
What is the third step of signaling control of gene expression in the transcription step?
Activation of a Transcription Factor
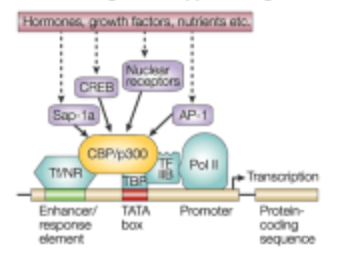
What is the fourth step of signaling control of gene expression in the transcription step?
Recruits other member of the transcription complex
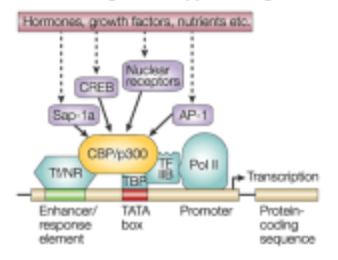
What is the fifth step of signaling control of gene expression in the transcription step?
Transcriptional Complex recruits RNA Polymerase II
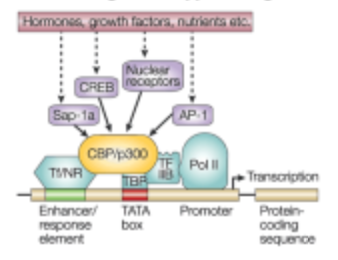
What is the sixth step of signaling control of gene expression in the transcription step?
Transcription is initiated at the promotor site.
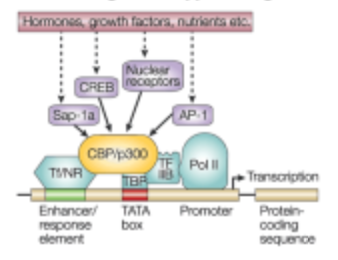
How is transcription turned on and off?
A transcription factor
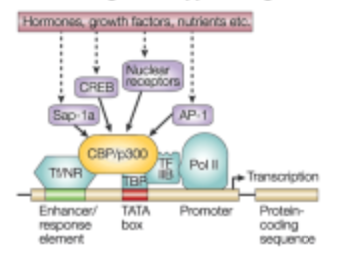
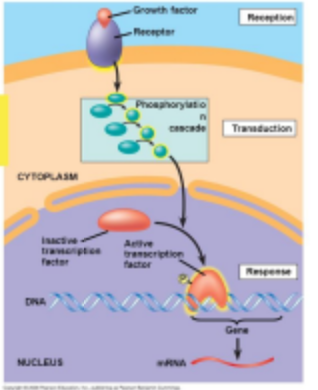
What can happen at any point in the following model?
A mutation that inhibits the results of the pathway.
What is the best way to stop gene expression?
As early as possible
What is a hormone?
A molecule that is prodiuced in one cellular location but whose effects are seen in another.
What do hormones require to have an effect?
A target cell with a receptor for the molecule, therefore no receptor means no response.
What is a transcription factor?
Some thing that controls the react of gene transcription either by helping or hindering RNA polymerase binding to DNA
What do transcription factors interact with other proteins to build?
Transcription complex that may increase transcription 100 fold.
What does a transcription complex contain?
A DNA Binding Domain that attaches specific DNA Sequences
What might a gene have for a distinct transcriptional factor?
Multiple binding sites, in addition a transcription factor might have a binding site on multiple different genes.
How can the amount of transcription instigated by a transcriptional factor be increased?
By an enhancer region of DNA and an activating protein somewhere in the vicinity of the gene that recruits the transcription machinery.
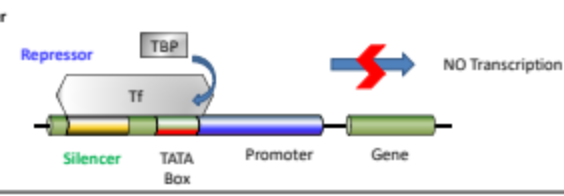
What is the TATA Box?
A DNA sequence found in the promotor region where transcription factor complex proteins bind, specifically TATA Binding Protein.
What is the Promotor?
A region of DNA located upstream of a gene that initiates transcription by providing a binding site for RNA polymerase and transcription factors.
What is the enhancer?
Region of DNA that Binds to Activator Proteins too activate the transcription process.
What is the silencer?
Region of DNA that binds to Repressor proteins to precent binding of RNA Pol II to the promote
What is the Insulator?
Region of DNA that blocks the interaction of enhancers with promoter using Insulator proteins. .
The core promotor is what?
The site of TATA binding protein and BASAL Factor binding; responsible for BASAL LEVEL OF EXPRESSION; Binding by GENERAL TF.
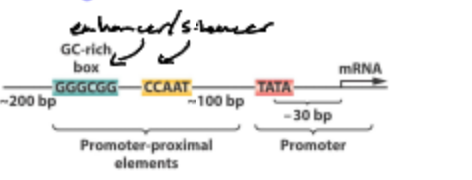
The Promoter Proximal Elements are what?
The site of additional ACTIVATOR PROTEINS; responsible for INDUCEDREPRESSED LEVEL OF EXPRESSION; Binding by TISSUE_SPECIFIC TF
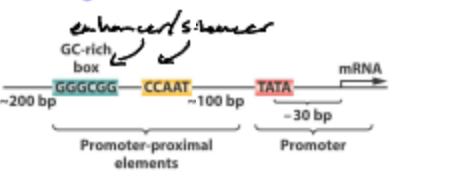
What does an enhancer do?
Supercharge transcription
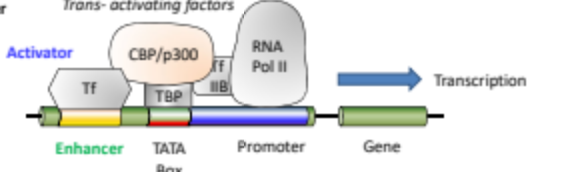
What does a silencer do?
Repress Transcription
Where can a Promotor-Proximal element be located?
Can lie far away from the gene up to 100kb away and even on the opposite DNA strand.
How does the Promotor-Proximal Region work?
Looping of DNA brings TF and Trans activating factors together leading to higher efficiency transcription of gene of interest.
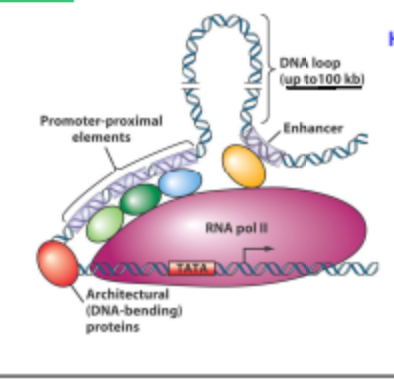
Is the insulator a cis or trans activating factor?
Both
What do insulators prevent?
The transcription of non target Genes
Since any mutation in transcription factors have the ability to be intimately associated with specific disease processes, what does that make transcription factors?
A prime target for potential drug discovery.
what is included in RNA Processing?
5’. Capping
#’ tail poly-denylation
RNA splicing
RNA transport
miRNA
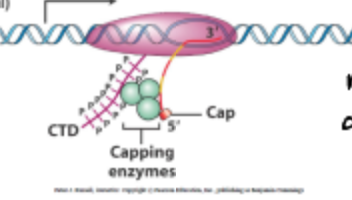
What is 5’ capping in relation to gene expression?
It is the addition of a modified guanine nucleotide to the 5' end of mRNA transcripts, which protects the mRNA from degradation and assists in ribosome recognition during translation.
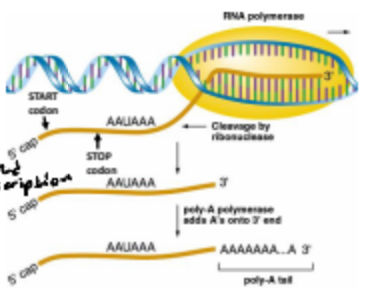
What is 3’ polyadenylation in regard to gene expression?
It is the addition of a poly-A tail to the 3' end of mRNA transcripts, which enhances mRNA stability, export from the nucleus, and translation efficiency.
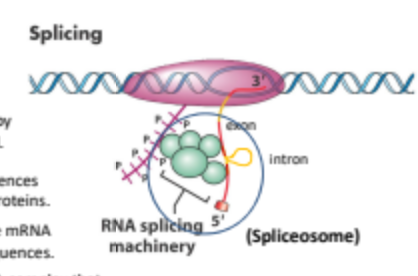
What is splicing in regar ot gene expression"?
Splicing is the process of removing introns from pre-mRNA and joining exons together to form a mature mRNA transcript that can be translated into a protein.
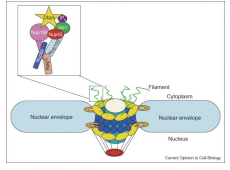
What is RNA Transport?
The export of mature MRNA from the nucleus to the cytoplasm controlled by a large number of Messenger Ribonuleaoproteins (mRNPs) MRNA export is through large multi protein pores complexes.
What protein is responsible for RNA transport through the nucleus?
mRNPs (Messenger Ribonucleoproteins)
What is mature microRNA?
miRNA is a class of naturally occurring, small noncoding RNA molecules whose main function is to down regulate gene expression. Typically around 20-25 nucleotides in length.
Why is miRNA highly useful as a biomarker?
It is deregulated in different types of cancer
What is the first step of miRNA regulation?
Transcription —> Processing
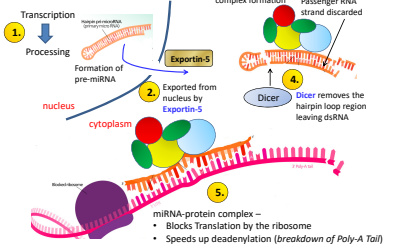
What is the second step of miRNA regulation?
Exported from nucleus by Exportin-5
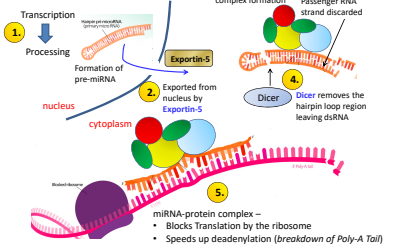
What is the third step of miRNA regulation?
miRNA Protein complex fomation and passenger RNA strand discarded.
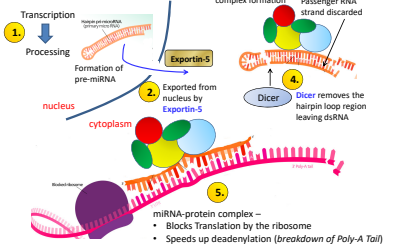
What is the fourth step of miRNA regulation?
Dicer removes the hair pin loop region leaving dsRNA
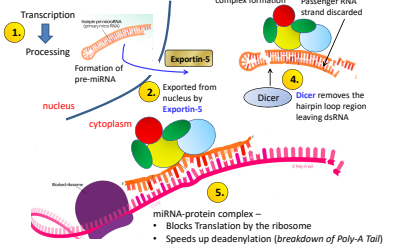
What is the fifth step of miRNA regulation?
miRNA protein complex blocs translation by the ribosome , speeding up deadenylation (breakdown of the Poly-A Tail)
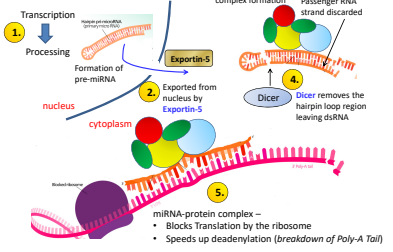
During transcription what is the first place that regulation can occur?
The promotor
During transcription what is the second place that regulation can occur?
5’ cap
During transcription what is the third place that regulation can occur?
Poly (A) tail
During transcription what is the fourth place that regulation can occur?
Protein Coding Sequence
During transcription what is the fifth place that regulation can occur?
Transport
During transcription what is the sixth place that regulation can occur?
miRNA
What is the most regulated step of translation?
Initiation
If there is a mutation with the 5’ cap what happens to the mRNA?
is likely to be degraded, preventing proper translation and subsequent protein synthesis.
If there is a mutation with the initiation codon what happens to the mRNA?
The mRNA cannot be properly processed or recognized by ribosomes, leading to its degradation and inhibiting translation.
If there is a mutation with the reading frame what happens to the mRNA?
The mRNA can produce incorrect amino acid sequences during translation, leading to dysfunctional proteins.
The elongation step of translation is dependent on elongation factors. What happens if there is a mutation with these factors?
The translation elongation process is impaired, resulting in stalled ribosomes and incomplete polypeptide chains.
What is Ribosomal Pausing?
Stacking of ribosomes on an mRNA molecules caused by changes in cellular environment. Can result in release of the ribosome and premature degradation of the incomplete polypeptide.
During Termination of Translation there is a protein called the release factor that causes termination at the stop codon. What happens if there is a mutation in the Release Factor?
The translation process fails to terminate correctly, leading to prolonged ribosome activity and potentially resulting in longer or nonfunctional polypeptide chains.
There are two groups of post translational regulation: Structural and Functional. What are the structural groups?
Disulfide Bonds, and Proteolytic Cleavage
What is proteolytic cleavage?
Cleavage of the pre-proteinsresulting in a mature fyunctional proteins.
i.e. Insulin
What if there is a mutation in proteolytic cleavage?
The maturation of proteins may be impaired, leading to the production of inactive or malfunctioning proteins, potentially disrupting normal cellular functions.
There are two groups of post translational regulation: Structural and Functional. What are the functional groups?
Myristoylation and phosphorylation
What is Myristoylation?
The attachment of lipid chains resulting in membrane localization of a protein.
What is phosphorylation?
The addition of a phosphate group to a protein, often regulating its activity or function.
What is up-regulation?
The process which results in an INCREASED expression of one or more genes.
What is down-regulation?
The process which results in DECREASED gene and corresponding protein expression.
Complete the following Genetic Material must-have:
Effective __________ between generations.
transmission of information
Complete the following Genetic Material must-have:
Ability to store _______ _________ ___ _________.
vast amounts of information
Complete the following Genetic Material must-have:
Information can be changed; _______
mutable
Complete the following Genetic Material must-have:
Effective __________ / High Fidelity.
Replication
If the amino acid in a peptide changes, then …
the configuration of the peptide may change.
What are the general types of point mutations?`
Substitutions, Insertions/Deletions, and Functional Mutants.
What is a transition mutant?
A substitution mutation where there is an exchange of a purine for a purine or a pyrimidine for a pyrimidine.
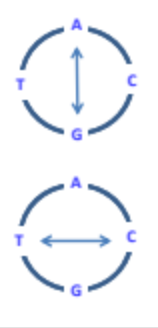
What is a transversion mutation?
A substitution mutation where there is an exchange of nucleotides outside the nucleotide family; exchange a purine for a pyrimidine.

What do substitution mutatiosn typically have an efffect on?
The differientiation between closely related species.
What happens more often; transitions or transversion mutations?
Transition because the structure of the compound does not have to change.
What are the three types of substitution mutations?
Silent Mutations, Missense Mutations, Nonsense Mutations
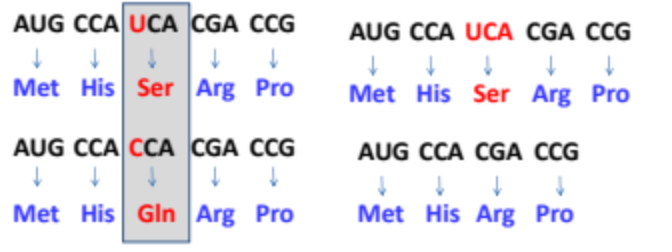
What is a silent mutation?
A mutation where changing one codon to a synonymous codon causes no change in the amino acid sequence of the protein. this illustrates the degeneracy of DNA
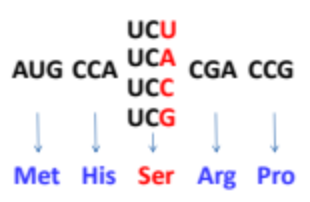
Where do silent mutations typically occur?
The third nucleotide in the codon.
What is a missense mutation?
The changing of one codon to a different codon resulting in a change in the amino acid sequence of the proteins. This type of mutation can lead to a nonfunctional protein or affect its function.
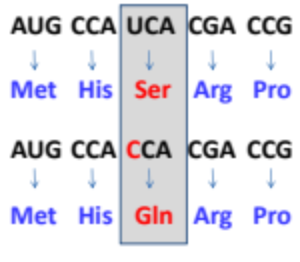
Why is the level of severity of a missense mutation rely on the position of the mutation?
changes in critical areas like active sites or structural domains can significantly impact protein function.
Where do missense mutations usually occur?
Almost always at the primary first position of the codon.
What is a nonsense mutation?
A change to the codon that results in a premature “STOP” codon resulting in a premature stoppage of the translation. Usually a very severe result.
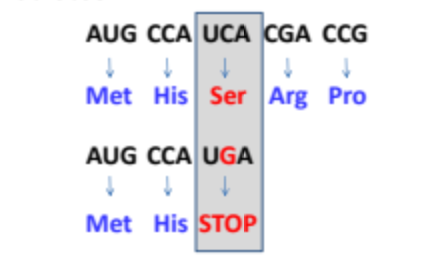
What can drugs like Translarna do for someone with a mutation?
They can potentially read through premature stop codons, allowing for the production of full-length proteins.
What are the two types of insertion/ deltion mutations
Frameshift mutations and in-frame mutations
What is a frameshift mutation?
A mutation that causes the gain or loss of a nucleotide (or nucleotides) that results in a change in the reading frame of the codon, thus effecting all codons there after.
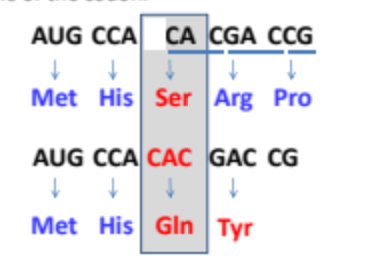
For insertion/deletion mutations what determines the severity of the mutation?
The position that the mutation occurs at.
True or False:
A frameshift muation can cause a premature stop codon?
True
What is an in-frame mutation?
A mutation that results in the gain or loss of a nucleotide or trinucleotide set that does not change the reading frame of the codon.