Biochemistry
1/67
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
68 Terms
exergonic/endergonic
energy exits/enters the system, negative/positive dG
exothermic/endothermic
heat exits/enters the system, negative/positive dH
entropy
dS is always positive, disorder of universe tends to increase
enthalpy
dH = dE + PdV, heat
Gibbs free energy
dG = dH - TdS, negative dG means reaction is spontaneous and favorable, this is determined by both Keq and Q
dG' = - RTlnK'eq
dG = dG' + RTlnQ, Q = Keq but not at any given time
ATP -> ADP + P, dG = -12
activation energy
energy required to produce the transition state, catalyst/enzyme stabilize the transition state and reduce Ea without changing dG
higher Ea means slower reaction rate
drawing a reaction coordinate graph
enzymes
physiological catalysts
increase reaction rate so it happens in a biologically relevant time-frame, not used up in reaction, specific to a reaction (important for regulation)
interact with substrate at active site, always stereospecific and can form specific stereoisomers from non-chiral molecules
can interact with different substrates that have similar chemical linkages
induced-fit model vs. lock-key model
dimers have two similar proteins connected by hydrophobic amino acids or by disulfide bonds
heterodimer- two different proteins
homodimer- two identical proteins
common types:
1. kinases takes phosphate group from donor (ATP)
2. phosphatases removes phosphate group
3. phosphorylases adds phosphate group
3. ligases combine two molecules
4. lyases break apart a molecule, form double bond
5. isomerases convert between isomers
6. transferases transfer functional groups from one molecule to another (sometimes includes kinases and phosphatases)
activating enzymes
zymogen is an inactive enzyme that needs to be cleaved
apoenzyme is an inactive enzyme that needs a cofactor
phosphorylation can activate/deactivate
allosteric interactions can regulate
hydrolyzing enzymes
hydrolysis breaks bonds
lipase- hydrolysis of lipids (triacylglycerol breaks apart into glycerol and 3 fatty acids)
protease- hydrolysis of proteins (proteins are cleaved to activate subunits)
endonuclease- hydrolysis of nucleotides in middle of a strand (restriction enzymes cut at palindromes)
exonuclease- hydrolysis of nucleotides at the ends of a strand
ribonuclease- hydrolysis of RNA (protected from my 5'-caps and 3'-poly A tails)
amylase, glycosidase- hydrolysis of carbohydrates
enzyme regulation
1. regulated at allosteric site
2. regulated by modifications like phosphorylation
on vs. off states
negative feedback- product inhibits enzyme
positive feedback- product activates enzyme
oxytocin is example of positive feedback, needs external regulator to eventually stop process
oxidation/reduction
loss/gain of hydrogen atoms, gain/loss of charge
Bronsted-Lowry acid/base
proton donor/acceptor
Lewis acid/base
electron pair acceptor/donor, usually in coordinate covalent bonds
acid/base-dissociation constant
large Ka/Kb means stronger acid/base
Ka = [H3O+][A-]/[HA]
Kb = [HB+][OH-]/[B]
amphoteric
can act as either acid or base, amino acids
conjugate base of a weak polyprotic acid is always amphoteric
each time a polyprotic acid donates another proton, it becomes a weaker acid
pH
pH = -log[H+], water at 25C has pH = 7
pH + pOH = 14
pKa
pKa = -logKa
lower pKa/pKb is the stronger the acid/base
buffer
weak acid and its conjugate base
bicarbonate buffer system, carbonic acid and bicarbonate
amino acids
memorize their structure, names, letters, properties, physiological pH
Nonpolar: PI GALVY MWF
"my PI goes to Galveston on mon/wed/fri"
Acidic: DE (negative at physiological pH)
Basic: HRK (positive at physiological pH)
alanine - ala - A
glycine - gly - G
valine - val - V
leucine - leu - L
isoleucine - ile - I
proline - pro - P
phenylalanine - phe - F
tryptophan - trp - W
tyrosine - tyr - Y (10.1)
serine - ser - S
threonine - thr - T
cysteine - cys - C (8)
methionine - met - M
lysine - lys - K (10.5)
arginine - arg - R (12.5)
histidine - his - H (6.1)
aspartic acid - asp - D (3.9)
glutamic acid - glu - E (4.1)
asparagine - asn - N
glutamine - gln - Q
amino group (10)
carboxyl group (2)
conservative substitution
binding affinity is not affected by the substitution, indicates that the original amino acid is not involved in binding or does not change conformation of enzyme
if binding affinity goes up or down, it is not conservative
average weight of amino acid
110 Da (g/mol)
Henderson-Hasselbalch equation
pH < pKa, will be protonated
pH > pKa, will be deprotonated
isoelectric point
pH of amino acid where net charge is 0, zwitterion
pI = average of the pKas of the two functional groups
peptide bond
amino group attacks the carboxyl group during synthesis
proteolytic cleavage breaks peptide bonds by hydrolysis
N-C synthesis, N-terminus is synthesized first and written first
disulfide bridge
cysteines are oxidized to cystine in a disulfide bridge
join together multiple subunits of proteins
reducing conditions in a gel will break apart disulfide bridges and thus the subunits
pi stacking
tryptophan and other aromatic compounds can undergo pi stacking interactions with each other
protein primary structure
order of amino acids, sequence, N-C synthesis, defined by peptide bonds
nonpolar sequences prefer to be on the inside of a protein or in the transmembrane region
protein secondary structure
alpha-helix is right-handed, 3.6 aa per turn, no prolines, favorable for transmembrane proteins, defined by hydrogen bonds between backbone components
beta-sheet is parallel or antiparallel, hydrogen bond
prolines and glycines are used for turns in protein structure
protein tertiary structure
interactions between residues in the chain
covalent:
disulfide bonds
non-covalent:
hydrophobic interactions- hydrophobic residues fold in the interior of the protein
polar interactions- van der Waals
ionic interactions- acid/base side groups
protein quarternary structure
interactions between residues between different polypeptides, allows connection of subunits to form protein
hydrophobic force
hydrophobic collapse- proteins fold to push hydrophilic sections to the exterior and hydrophobic sections to the interior
solvation shell- water molecules interact unfavorably with hydrophobic sections, so water molecules forced to lock their orientation and form shells around the protein
this low entropy and high free energy state is relieved by protein folding
protein unfolding
sigmoidal since its a cooperative process
determines thermodynamic stability
drugs
IC50 is concentration of drug that inhibits 50% of cells
Kd is dissociation constant between drug and target, with a lower value indicating higher affinity
drugs are metabolized by the body for excretion
vmax
all active sites on enzymes are occupied, vmax constant
kinetic measurement
depends on:
1. type of enzyme
2. concentration of enzyme
when there is high concentration of substrate. vmax is more important than Km
Km
concentration of substrate to reach 1/2 vmax
thermodynamic measurement
essentially the affinity of E for S, high affinity means low Km, depends on the properties of the binding site
always assume reversibility
if vmax is lowered than Km is lowered too
when there is low concentration of substrate, Km is more important than vmax
competitive inhibition
same vmax, increased km
inhibitor binds at active site
can always be out competed by additional substrate, so vmax doesn't change
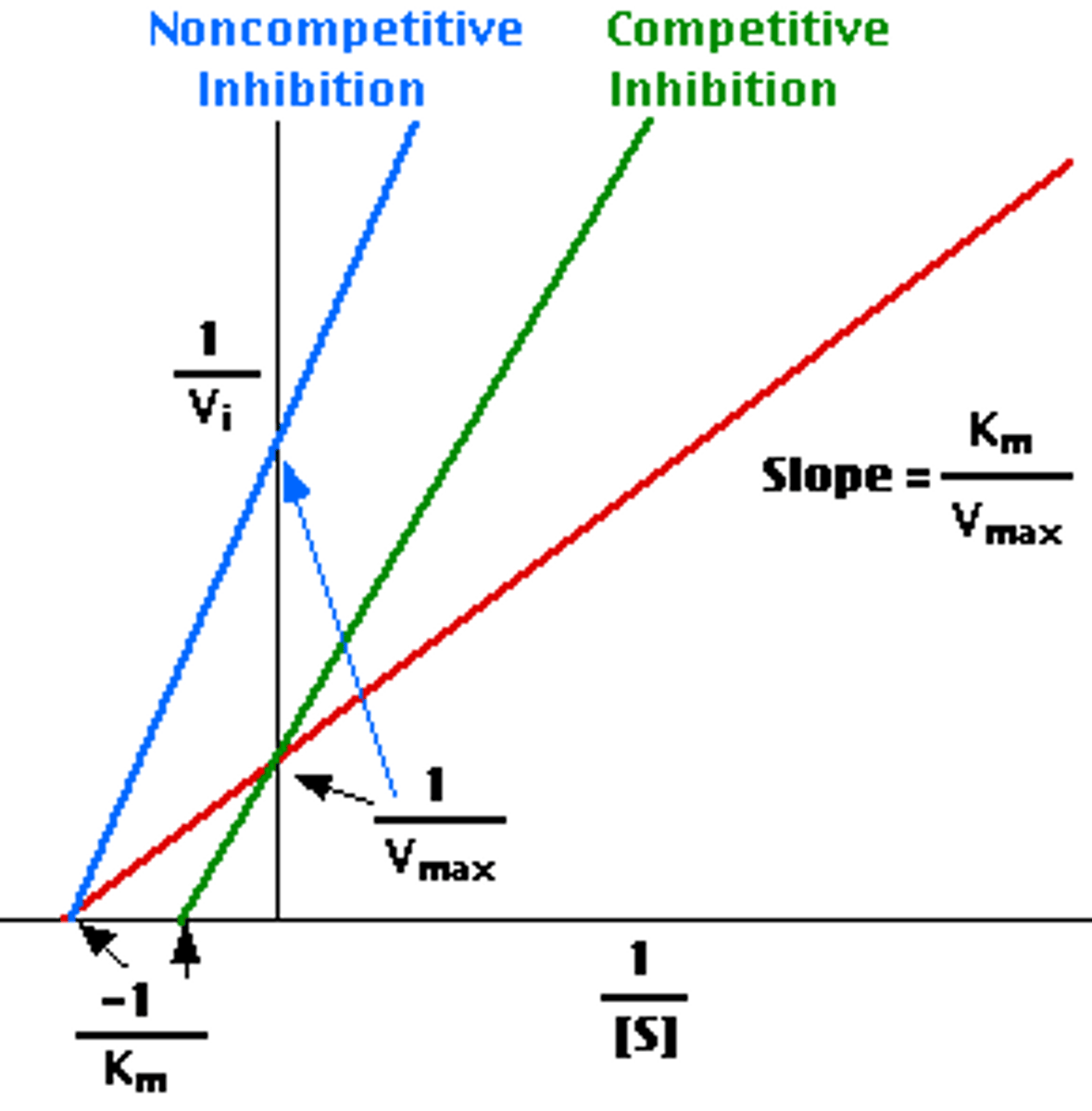
noncompetitive inhibition
decreased vmax, same km
inhibitor binds at allosteric site
alters shape of active site, so vmax decreases
binds to enzyme and enzyme-substrate substrate with same affinity
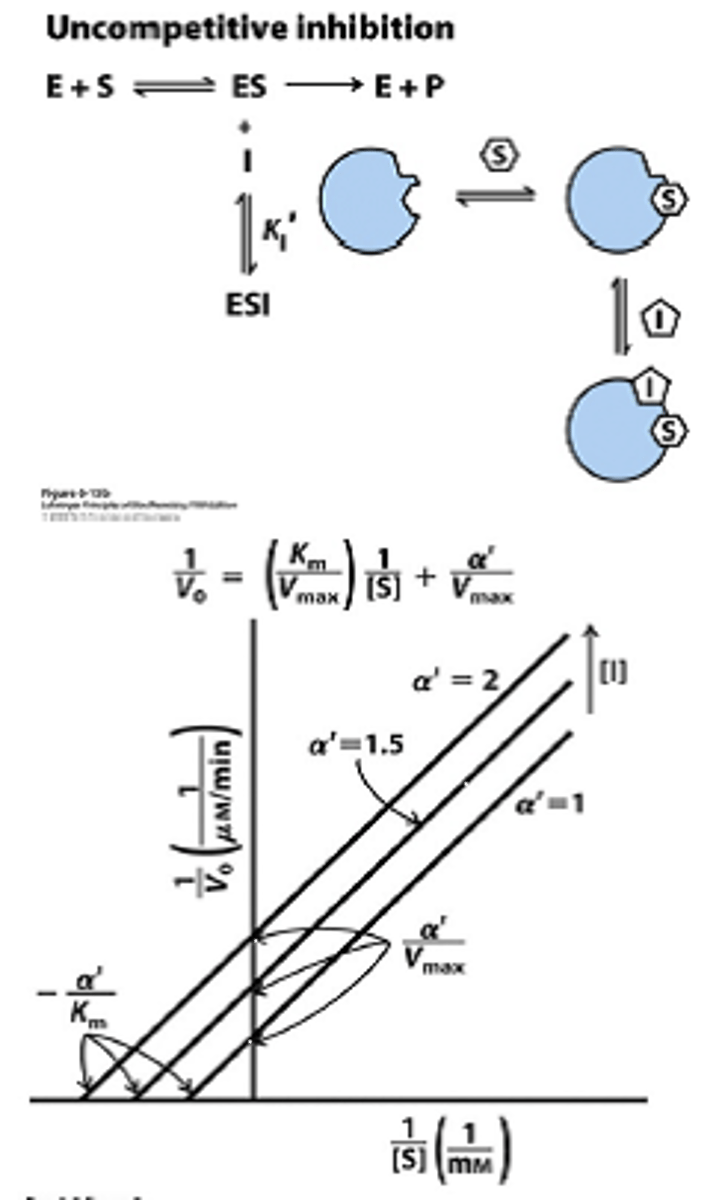
uncompetitive inhibition
decreased vmax, decreased Km
inhibitors binds to ES complex, mixed inhibition type I
increased substrate increases inhibitor effectiveness
locks S in active site, so decreases vmax and km
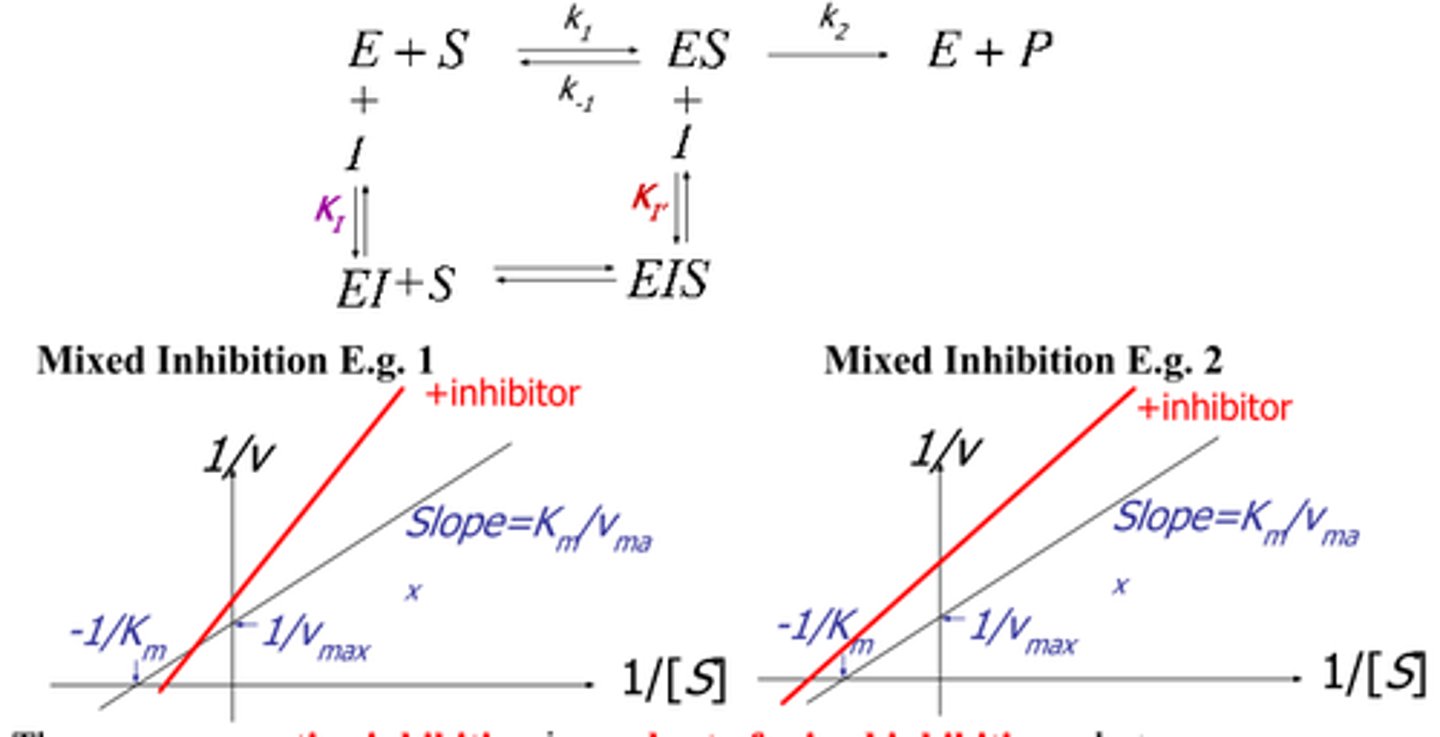
mixed inhibition
decreased vmax, increased/decreased km
binds at allosteric site, better affinity than substrate
type I: prefers to bind ES complex, so decreased Km
type II: prefers to bind E alone, so increased Km
Michaelis-Menten equation
v = vmax[S]/(Km+[S])
vmax = k_cat*[E]
catalytic efficiency = k_cat/Km
![<p>v = vmax[S]/(Km+[S])</p><p>vmax = k_cat*[E]</p><p>catalytic efficiency = k_cat/Km</p>](https://knowt-user-attachments.s3.amazonaws.com/60a54172-7b15-4d68-8d85-2a025ca760fc.jpg)
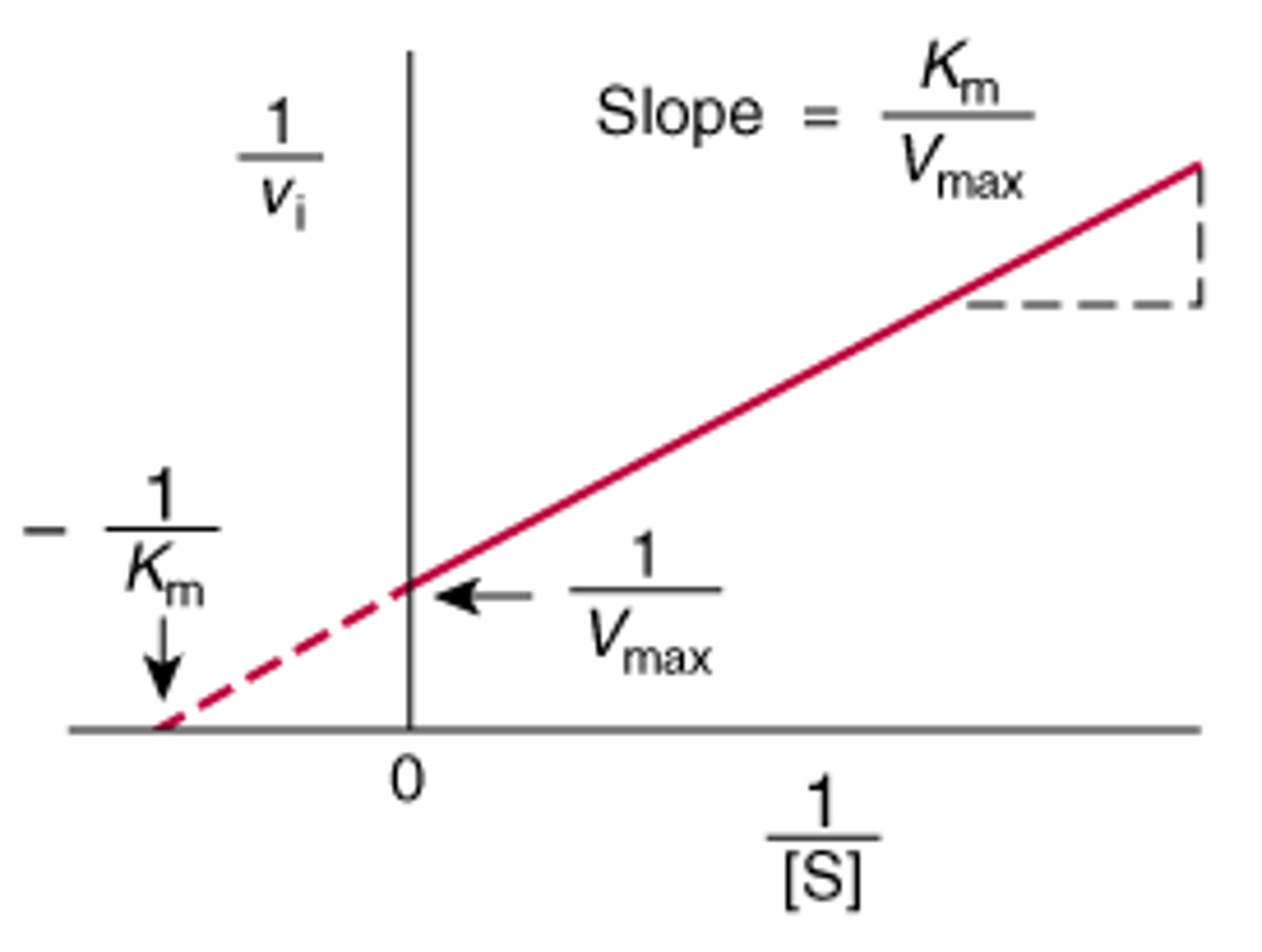
Lineweaver-Burke plot
inverse of rxn speed is y
inverse of substrate conc. is x
1/Vmax is y-intercept
1/Km is x-intercept
useful for testing inhibitor's effect on vmax and Km, plot two lines with and without inhibitor
slope is Km/vmax
ternary complex mechanisms
both substrates occupy active site at same time:
1. ordered mechanism- one substrate must bind first
2. random order mechanism- doesn't matter which is first
specific activity
units of enzyme per total protein mg
use specific activity to calculate purity
use just the units of enzyme (specific activity times total protein) to calculate yield
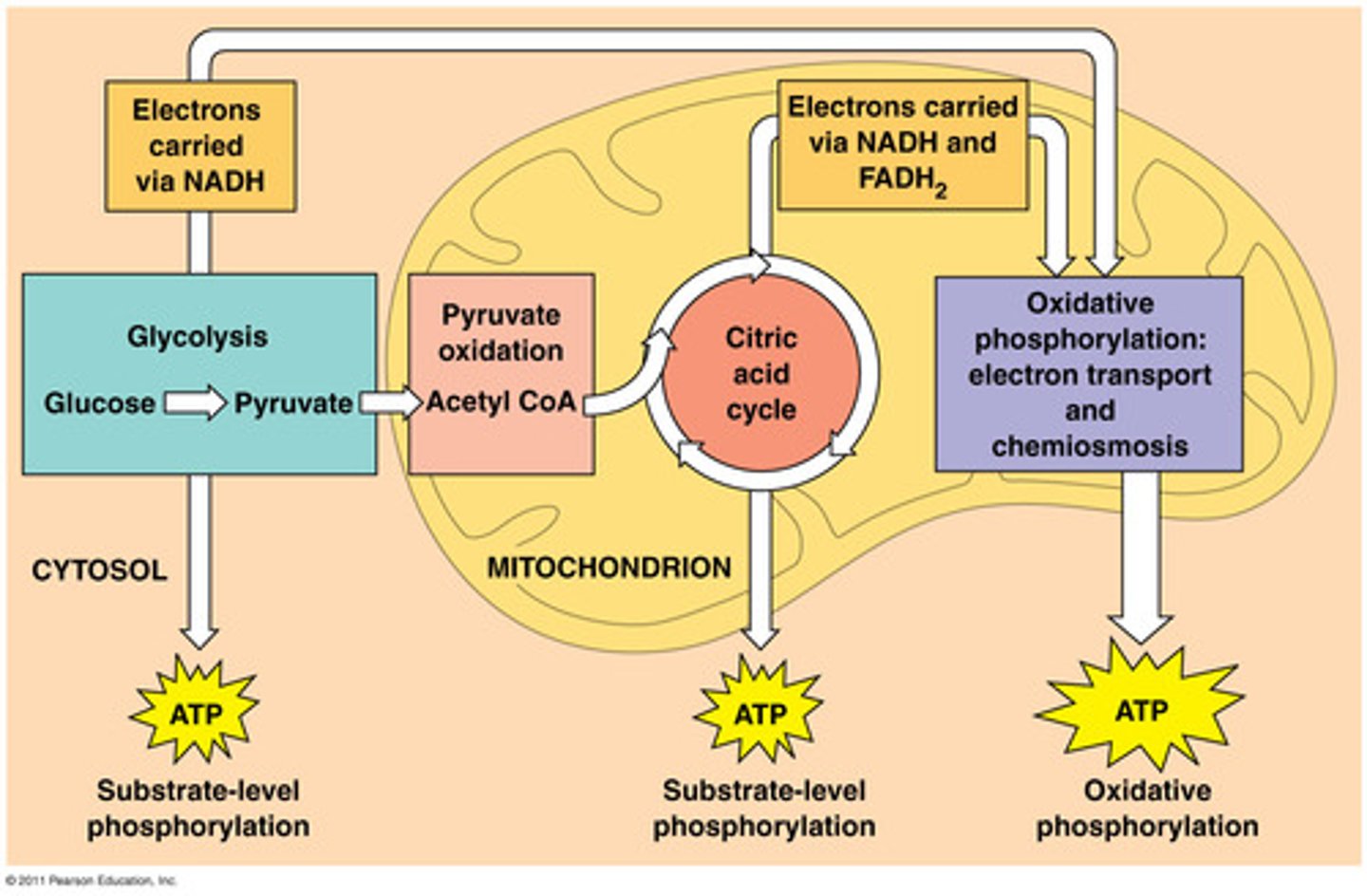
cellular respiration
NAD (nicotinamide adenine dinucleotide) and FAD (flavin adenine dinucleotide) accept electrons by getting reduced, later get oxidized on delivery to ETC
glucose is oxidized to CO2, O2 is reduced to H2O
1. glycolysis occurs in the cytosol
2. PDC and TCA cycle occurs in mitocondrial matrix, except in prokaryotes where it occurs in cytosol
3. ETC and oxidative phosphoylation occurs on inner mitochondrial membrane, except in prokaryotes where it occurs on cell membrane
electron carriers
NADH -> NAD+ + H+ + 2e-
FADH2 -> FAD+ + H+ + 2e-
NADH and FADH2 carry 2 electrons
CoQ carries 1 or 2 electrons
cytochrome C carries 1 electron
glycolysis
all cells possess this pathway, occurs in cytoplasm
glucose is oxidized and split into two pyruvates, produced net 2 ATP, 2 NADH
3 key steps:
1. hexokinase- converts glucose to glucose-6-P, uses ATP
2. phosphofructokinase (PFK) - converts fructose-6-P to fructose-1,6-P2, committed step of glycolysis, allosterically inhibited by high ATP
3. pyruvate kinase- converts PEP to pyruvate, produces 2 ATP
"goodness gracious, father franklin did go buy phat pumpkins (to) prepare pies"
"GG, final fantasy did get boring playing people punching people"
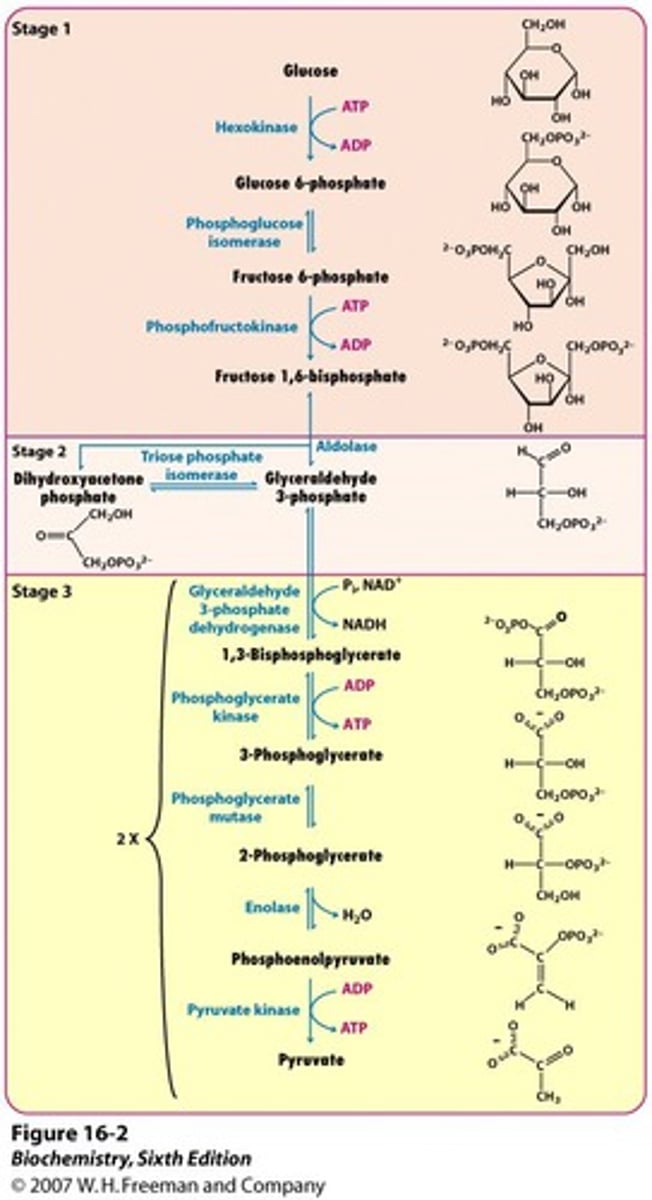
steps in glycolysis that create/require energy
require ATP (2 ATP investment for 1 glucose):
1. hexokinase- glucose to glucose-6-P
2. PFK- fructose-6-P to fructose-1,6-P2
create ATP (4 ATP total, 2 ATP net for 1 glucose):
1. phosphoglycerate kinase- 1,3-bisphosphoglycerate to 3-phosphoglycerate
2. pyruvate kinase- PEP to pyruvate
create NADH (2 total for 1 glucose):
1. GAP DH- glyceraldehyde-3-P to 1,3-bisphosphoglycerate
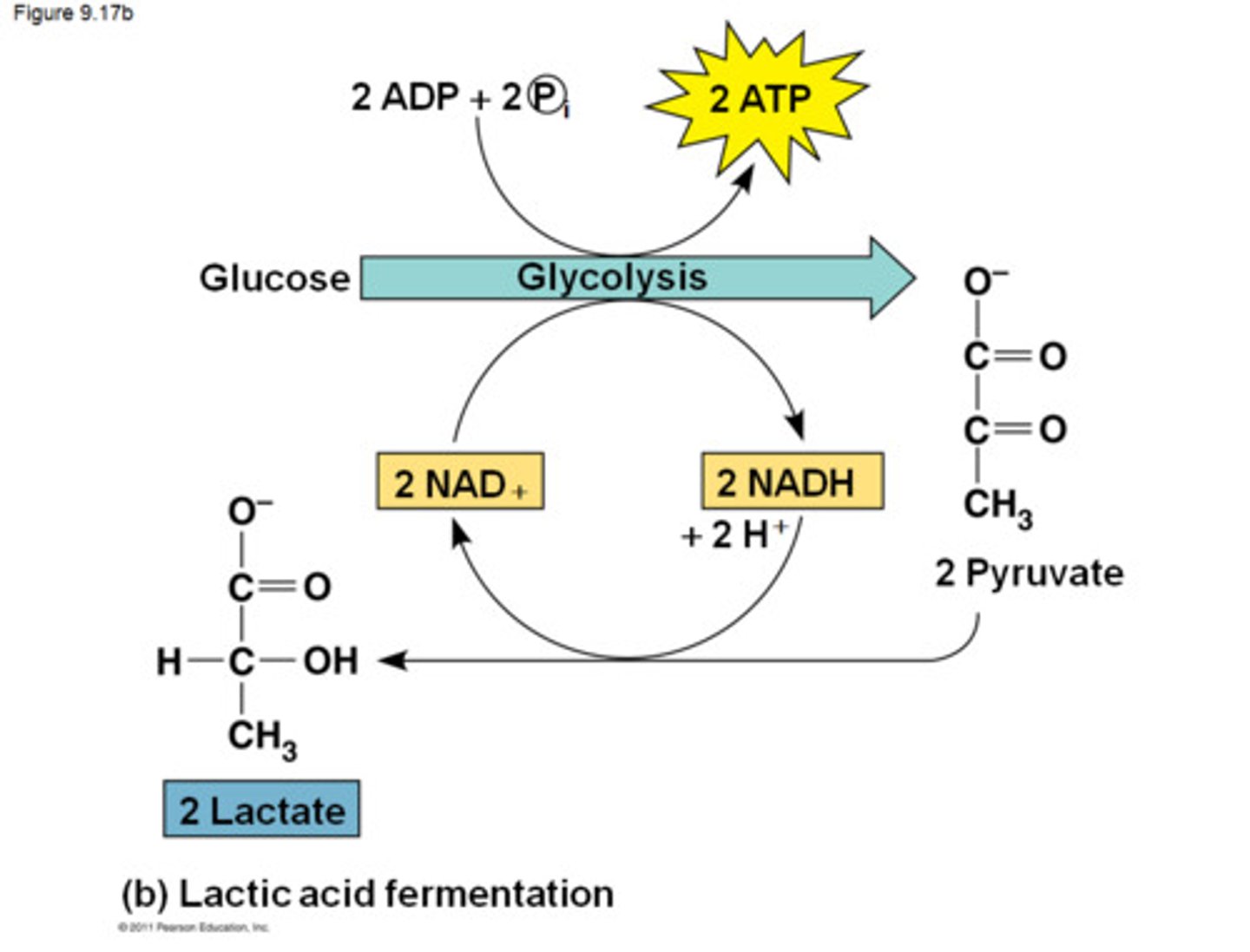
fermentation
aerobic conditions- pyruvate enters Krebs cycle, NADH from glycolysis is oxidized in ETC
anaerobic conditions- 2 ATP produced, 2 NADH must go back to regenerate NAD+ to continue glycolysis
to regenerate NAD+, pyruvate reduced to ethanol (yeast) or lactate (muscle), toxic when building up
pyruvate dehydrogenase complex
oxidative decarboxylation, pyruvate oxidized to acetyl-CoA (loses a carbon)
uses up CoA, NAD+ reduced to NADH, releases CO2
TPP- prosthetic group which is a covalently bound cofactor that helps with decarboxylation, derived from thiamine (vitamin B)
thiamine deficiency would increase rate of anaerobic glycolysis
allosteric regulation- ATP and fatty acids inhibit, since acetyl-CoA goes to fatty acid synthesis and ATP synthesis
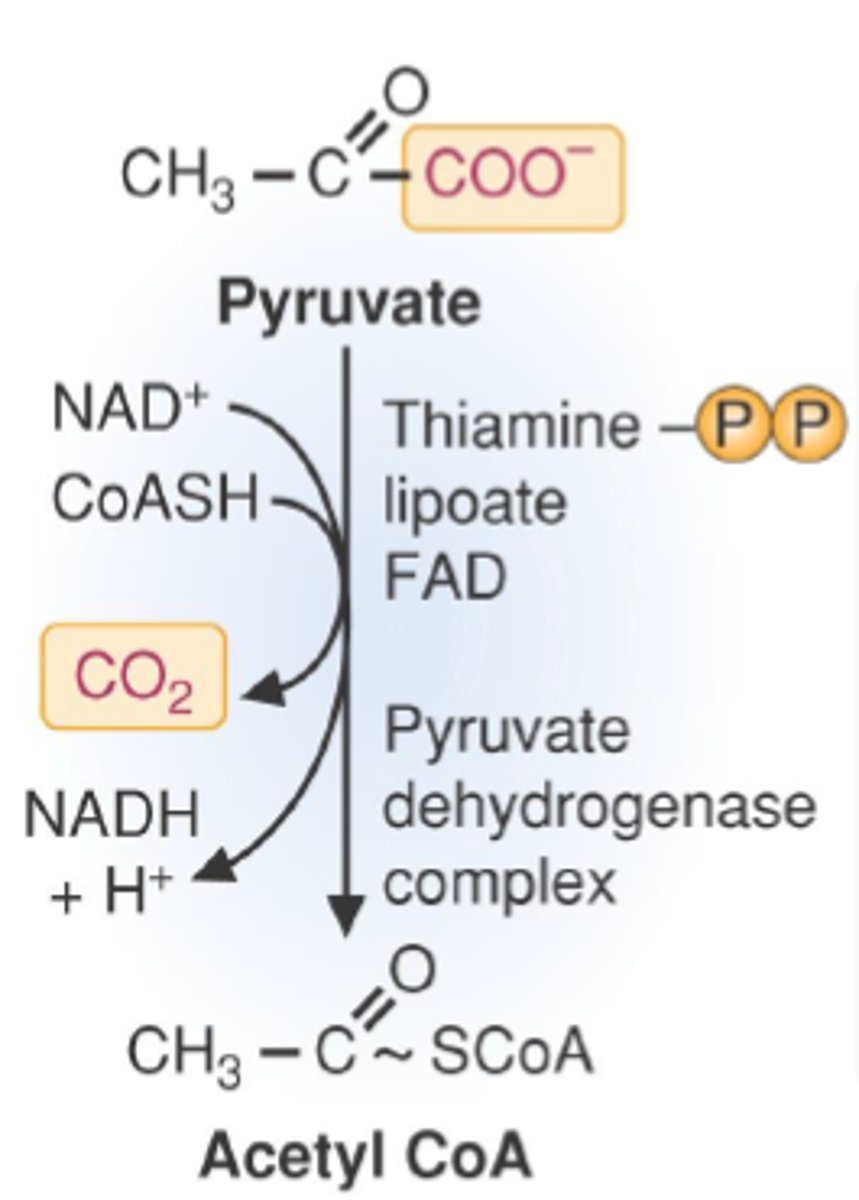
cofactors of pyruvate dehydrogenase complex
1. TPP- thiamine derived
2. lipoic acid
3. FAD+
4. NAD+ (converted to NADH, so technically not cofactor)
5. CoASH (attached to pyruvate to form acetyl-CoA)
TCA cycle
acetyl-CoA converted to citric acid, OAA from previous cycle also converted to citric acid
each turn produces 2 CO2, 3 NADH, 1 GTP, 1 FADH2
each glucose does two turns
aconitase- only enzyme name that doesn't match product
"can I keep selling sex for money, officer?"
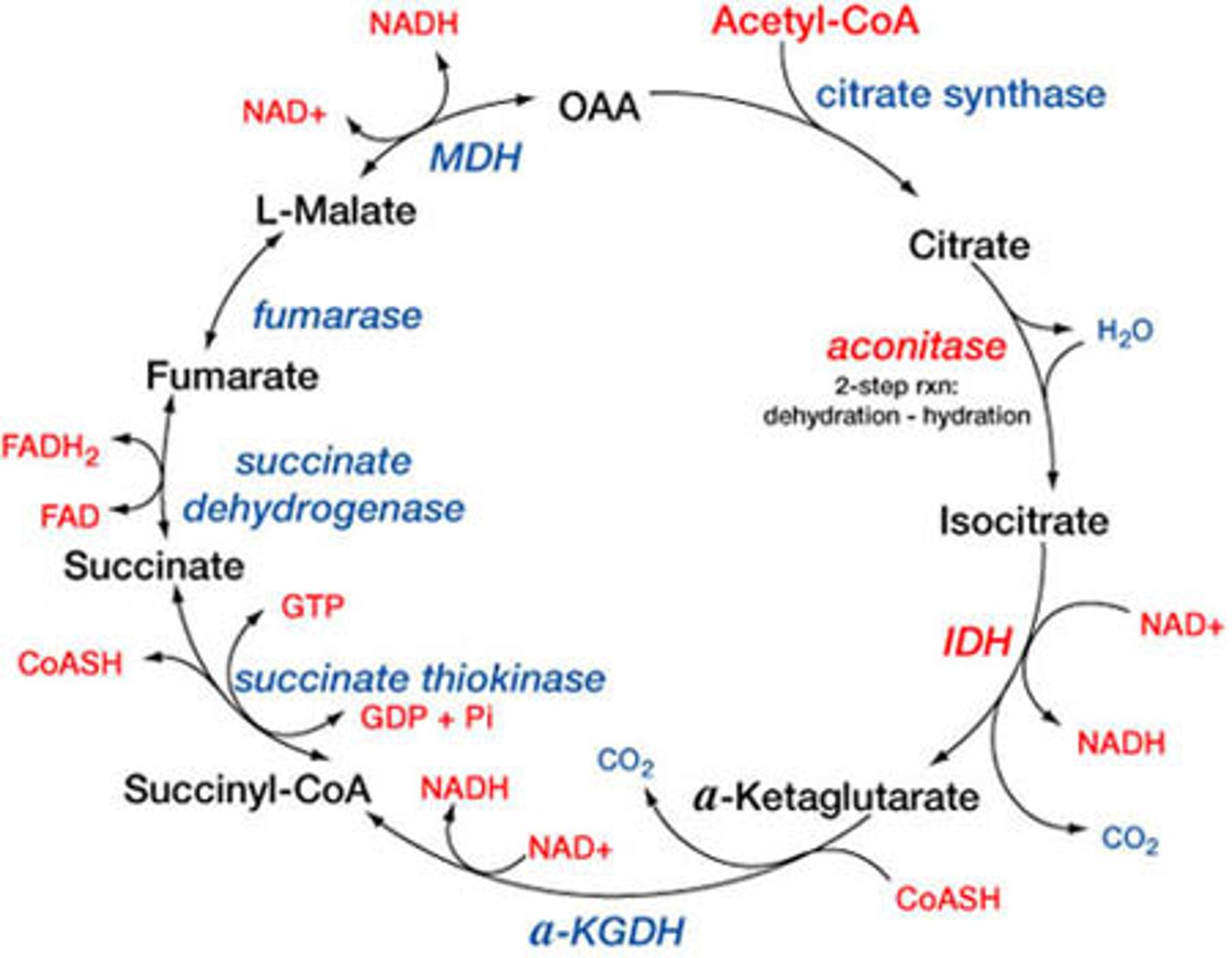
regulation of TCA cycle
substrate availability- amino acids can be converted to alpha-ketoglutarate to speed up TCA cycle
substrates inhibit their own enzyme- citrate inhibits citrate synthase, succinyl-CoA inhibits aKG DH
allosteric regulation- ATP, NADH inhibit TCA cycle
steps in TCA cycle that create energy
create NADH (3 total for 1 turn, 6 total for 1 glucose):
1. pyruvate DH complex- pyruvate to acetyl-CoA (technically not part of TCA cycle)
2 isocitrate DH- isocitrate to alpha-ketoglutarate
3. aKG DH- alpha-ketoglutarate to succinyl-CoA
4. malate DH- malate to OAA
create GTP (1 total for 1 turn, 2 total for 1 glucose):
1. succinyl-CoA synthetase- succinyl-CoA to succinate
create FADH2 (1 total for 1 turn, 2 total for 1 glucose):
1. succinate DH- succinate to fumarate
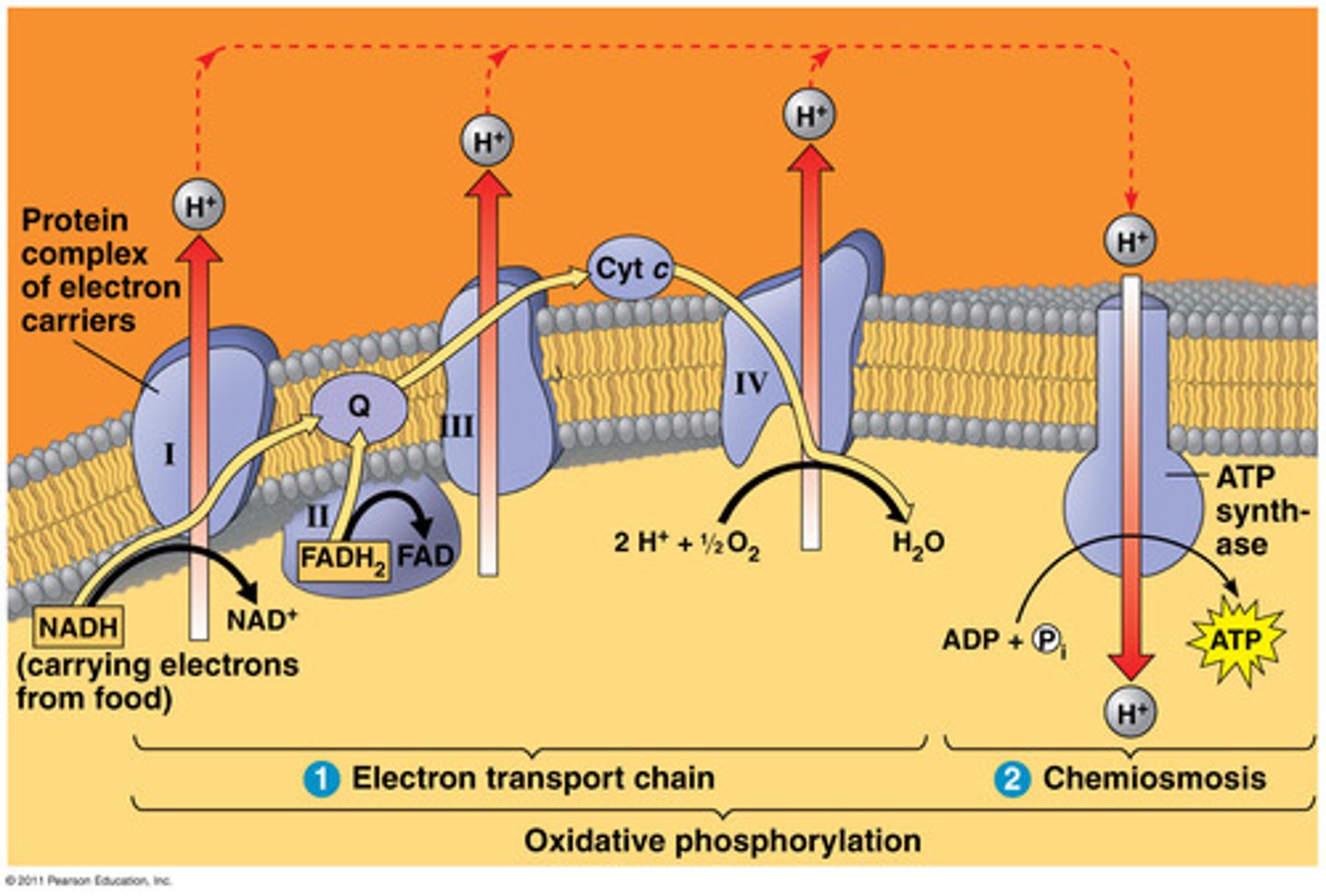
oxidative phosphorylation
two steps:
1. ETC- empty the electron carriers
2. chemiosmosis- make ATP
3 complexes pump H+ to intermembrane space:
1. NADH dehydrogenase- converts NADH to NAD+, CoQ carries electrons to complex 3
2. converts FADH2 to FAD+, CoQ carries electrons to complex 3
3. cytC reductase- cytC carries electrons to complex 4
4. cytC oxidase- O2 accepts electrons, converts to H2O
ATP synthase- H+ flows allowed to flow from intermembrane space to matrix, converts ADP to ATP
NADH produces 3 (2.5) ATP, moves 10 H+
FADH2 produces 2 (1.5) ATP, moves 6 H+
energetics of glucose catabolism (ATP count)
1. glycolysis- 2 ATP, 2 NADH (5 - 2 to bring NADH into mitochondria = ~3 ATP)
2. PDC- 2 NADH (~5 ATP)
3. 2 GTP (2 ATP), 6 NADH (~15 ATP), 2 FADH2 (~3 ATP)
ideal total: 38 ATP per glucose (actual: 30 ATP)
prokaryotes ideal total: 38 ATP
anaerobic glycolysis: 2 ATP
gluconeogenesis
activated by low glucose, high ATP
requires 6 ATP, 2 NADH to convert pyruvate to glucose
slightly different from glycolysis because pyruvate kinase is irreversible, so instead pyruvate is converted to OAA, then to PEP
bypasses acetyl-CoA, which means fatty acids cannot be converted to glucose
first step by pyruvate carboxylase happens in mitochondria, then transported out to cytosol
formation of glucose, fructose-6-P, and PEP are irreversible steps that push equilibrium to favor gluconeogenesis
glycogen- stored in liver, converted to glucose
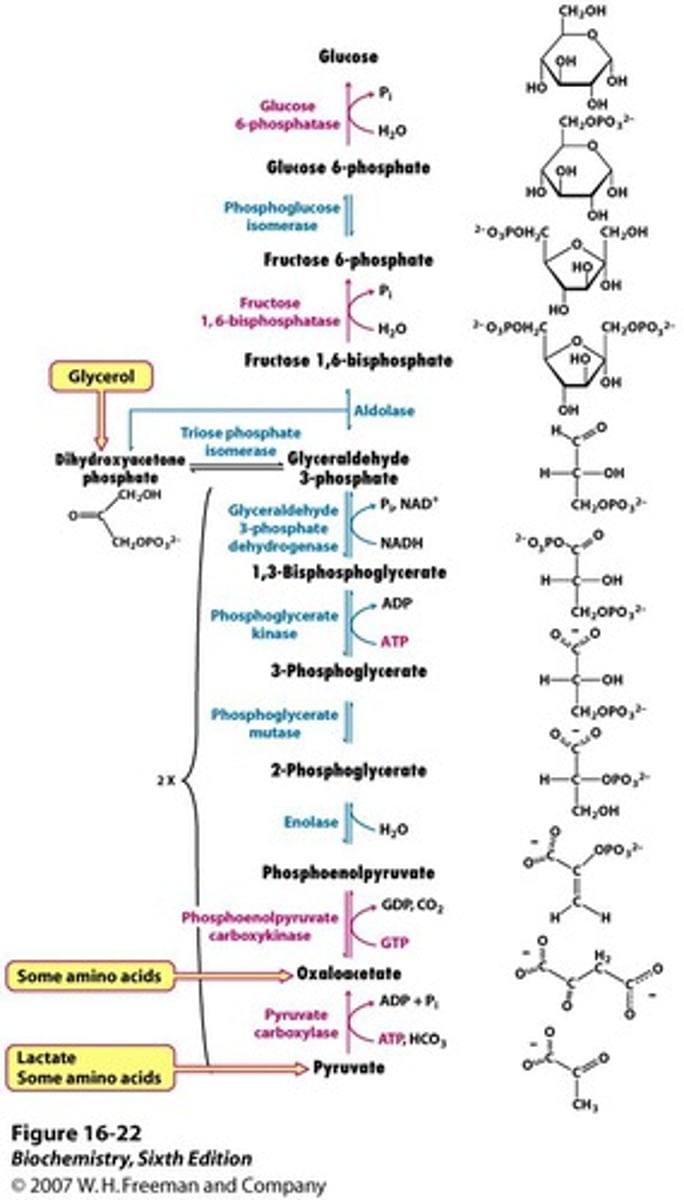
steps in gluconeogenesis that require energy
require ATP (6 total for 1 glucose):
pyruvate carboxylase- pyruvate to OAA
PEP carboxykinase- OAA to PEP
phosphoglycerate kinase- 3-phosphoglycerate to 1,3-bisphosphoglycerate
require NADH (2 total for 1 glucose):
GAPDH- 1,3-bisphosphoglycerate to glyceraldehyde-3-P
adding phosphate to glucose and fructose at the end does not require ATP
starting materials of gluconeogenesis
lactate, pyruvate, glycerol (enters through DHAP), amino acids (enter through pyruvate), any TCA cycle intermediates (enter through OAA)
glycogenolysis
glycogen is converted to glucose-6P
glycogen phosphorylase- just add phosphate group, no ATP required
regulation:
allosteric- ATP and glucose inhibit glycogenolysis
hormonal- epinephrine and glucagon activates glycogenolysis, insulin inhibits, all done through cAMP/pkA signalling
3 endpoints:
1. glycolysis- energy source for muscles
2. gluconeogenesis- glucose-6-phosphatase is only in live, converts to glucose and releases to blood
3. pentose phosphate pathway
regulation of cellular respiration
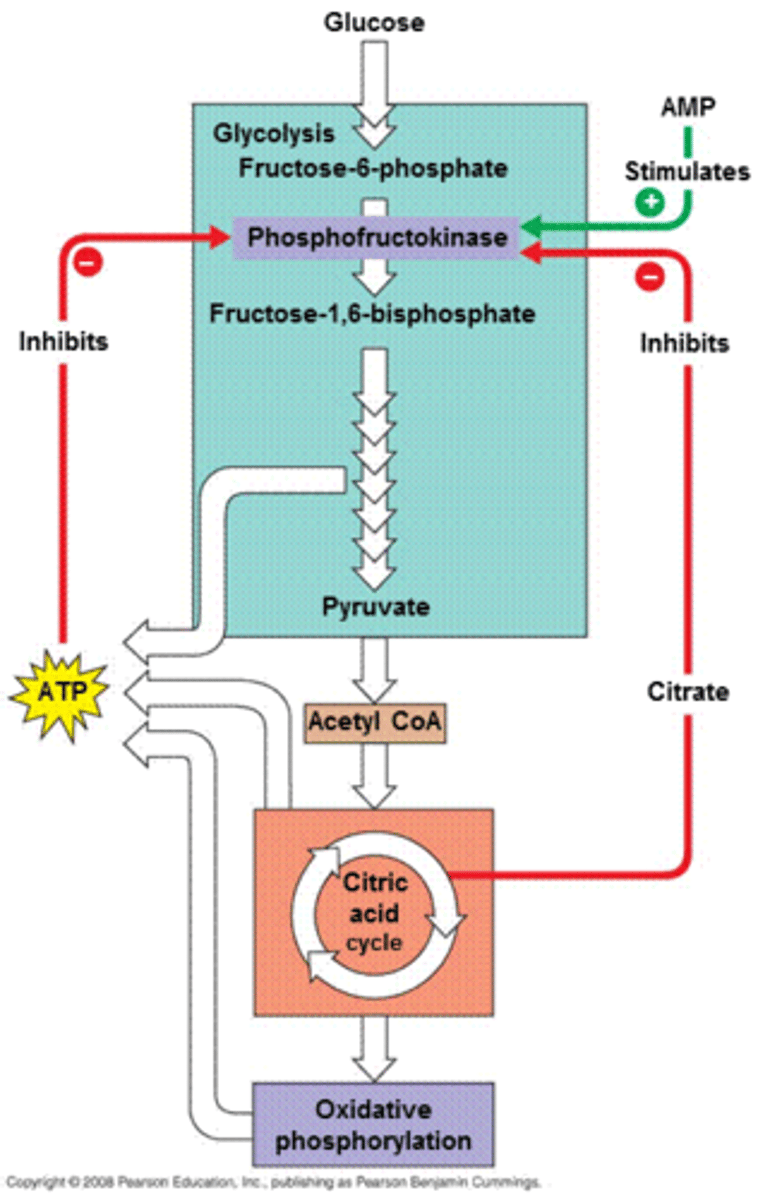
high ATP and citrate indicate Kreb's cycle activity, both inhibit PFK allosterically, activate fructose-1,6-P2ase (FBP)
substrate availability- glucose influx activate glycolysis, OAA influx activates gluconeogenesis
insulin- released with high glucose, activate PFK, promotes glycolysis, also recruits glucose transporters to plasma membrane, increases storage in glycogen and lipids
glucagon- released with low glucose, inhibit PFK, activate FBP, promotes gluconeogenesis, breaks down stored glycogen and lipids
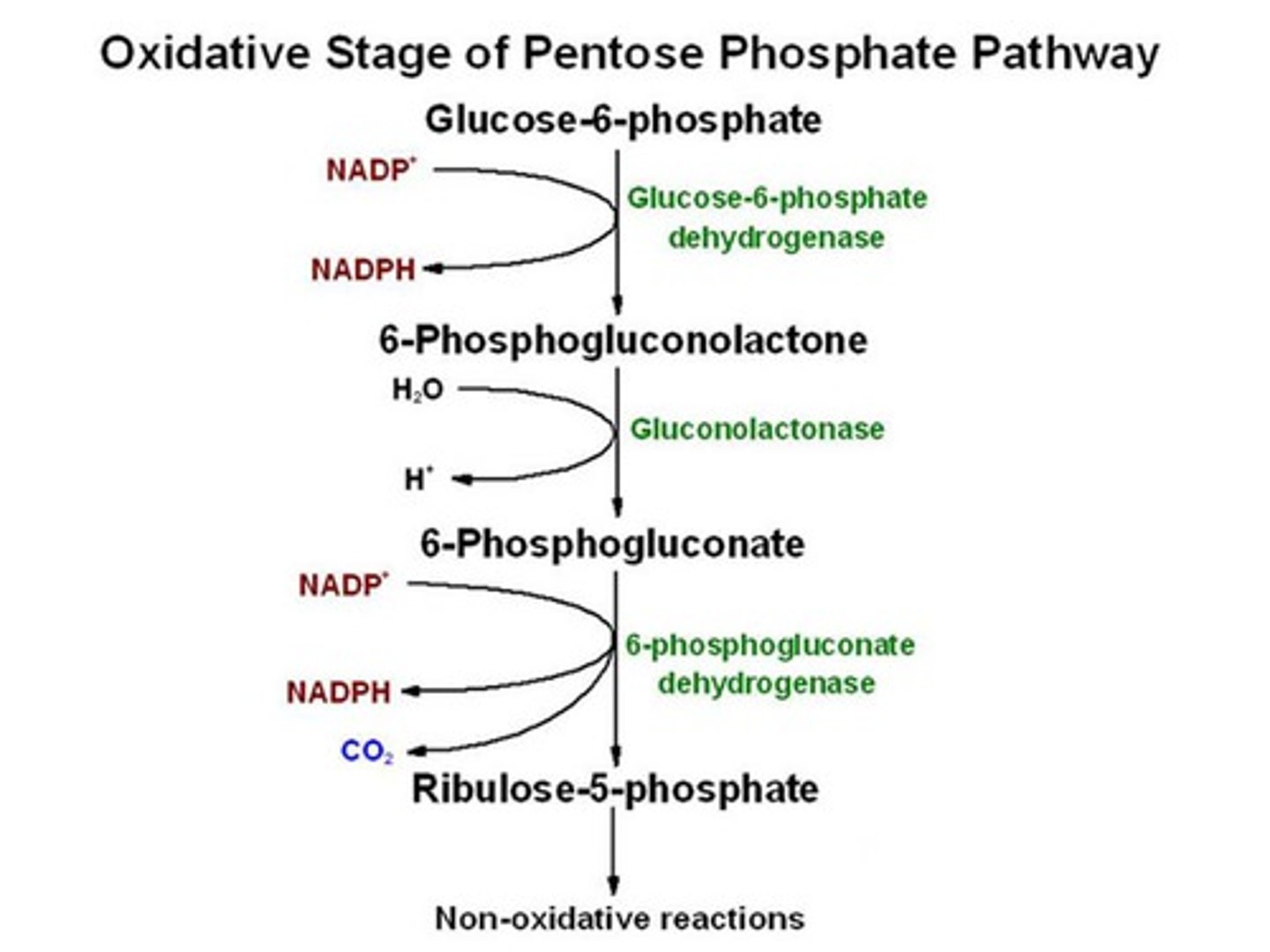
pentose-phosphate pathway
starts with glucose-6-P getting converted by GAPDH
releases 2 NADPHs total
oxidative phase- glucose-6-P converted to ribulose-5-P, 2 NADPHs and CO2 produced
non-oxidative phase- ribulose-5-P converted to ribose-5-P and glycolysis intermediates (2 GAP, 2 fructose-6-P)
it takes 3 glucose-6-P to make it through both phases
3 goals:
1. NADPH for reducing power in fatty acid synthesis
2. NADPH for eliminating free radicals
3. ribose-5-P for producing nucleotides
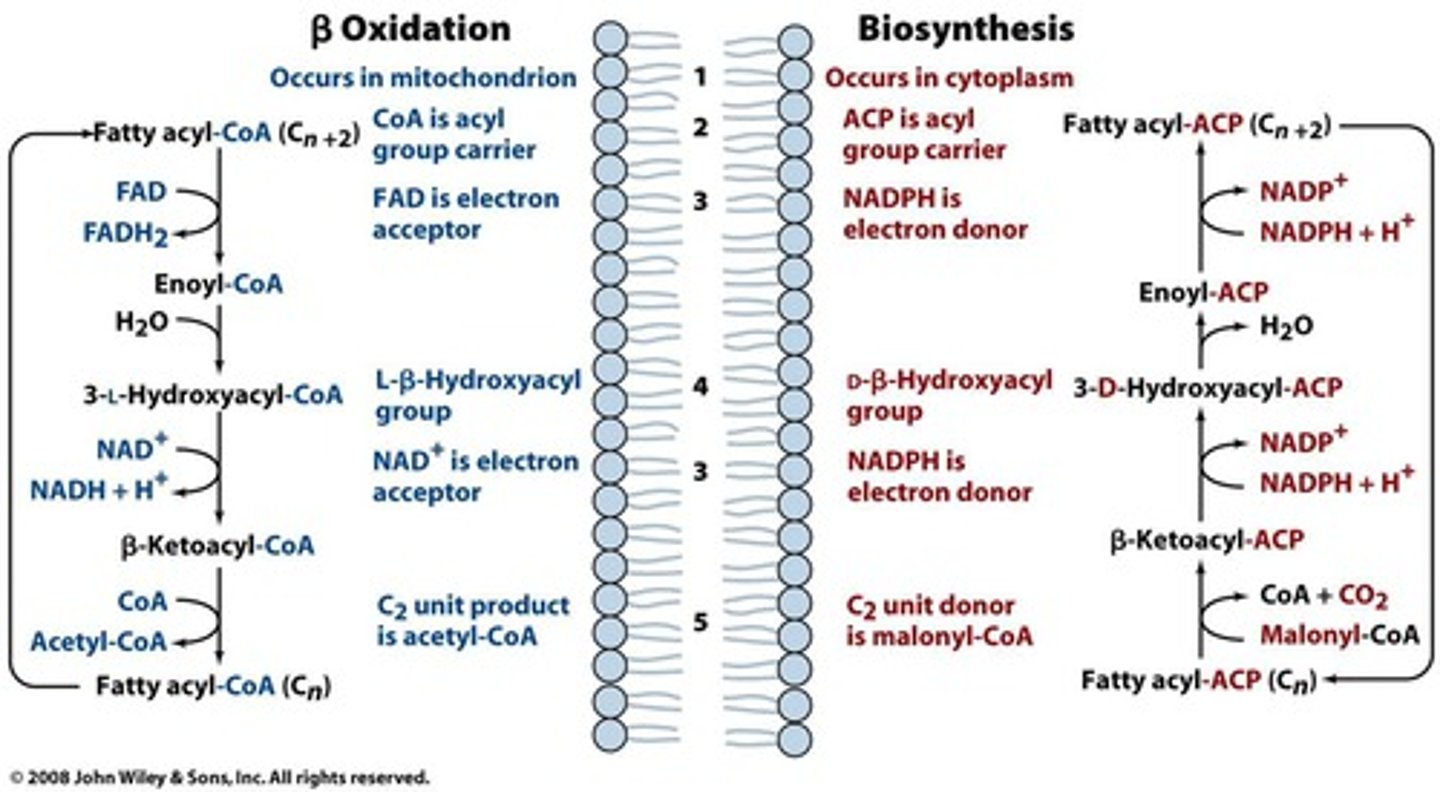
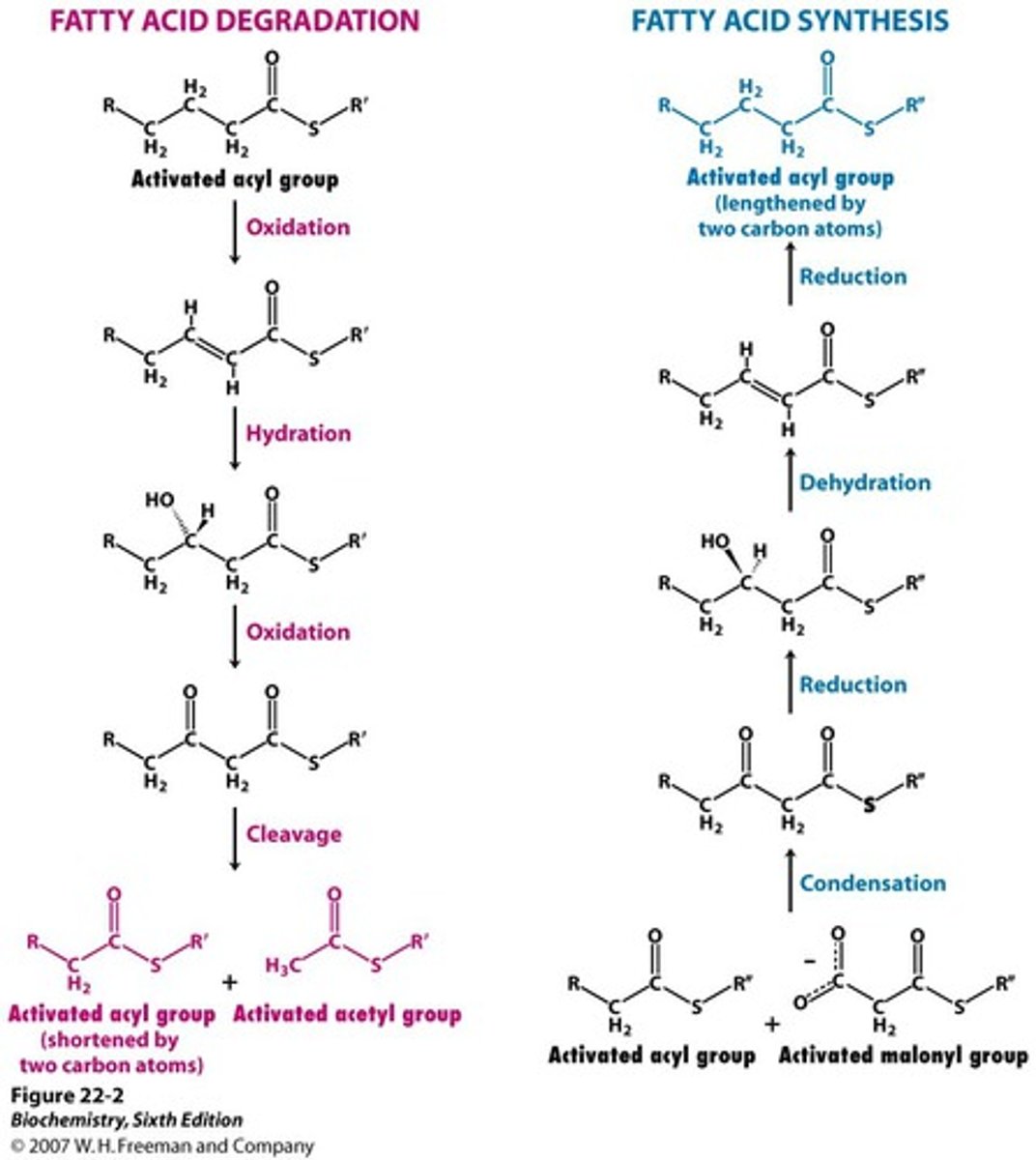
fatty acid oxidation (beta oxidation)
saturated fatty acids- dehydrogenase to create double bond (produce FADH2), then produce NADH to create ketone, breaks off acetyl-CoA, repeat
unsaturated fatty acids- isomerase to move double bond, then produce NADH to create ketone, break off acetyl-CoA, repeat
in mitochondrial matrix
needs 2 ATP to initially activate fatty acid
need 1 FAD, 1 NAD+ for each 2 C removed
produces 1 FADH2, 1 NADH
fatty acid metabolism
energy stored as triglycerides, glycerol, fatty acids
lipase is enzyme that breaks down triglycerides
1. in cytosol, fatty acid activated by addition of S-CoA to carboxylic end
2. in matrix, fatty acid undergoes beta oxidation to acetyl-CoA
3. goes to TCA cycle
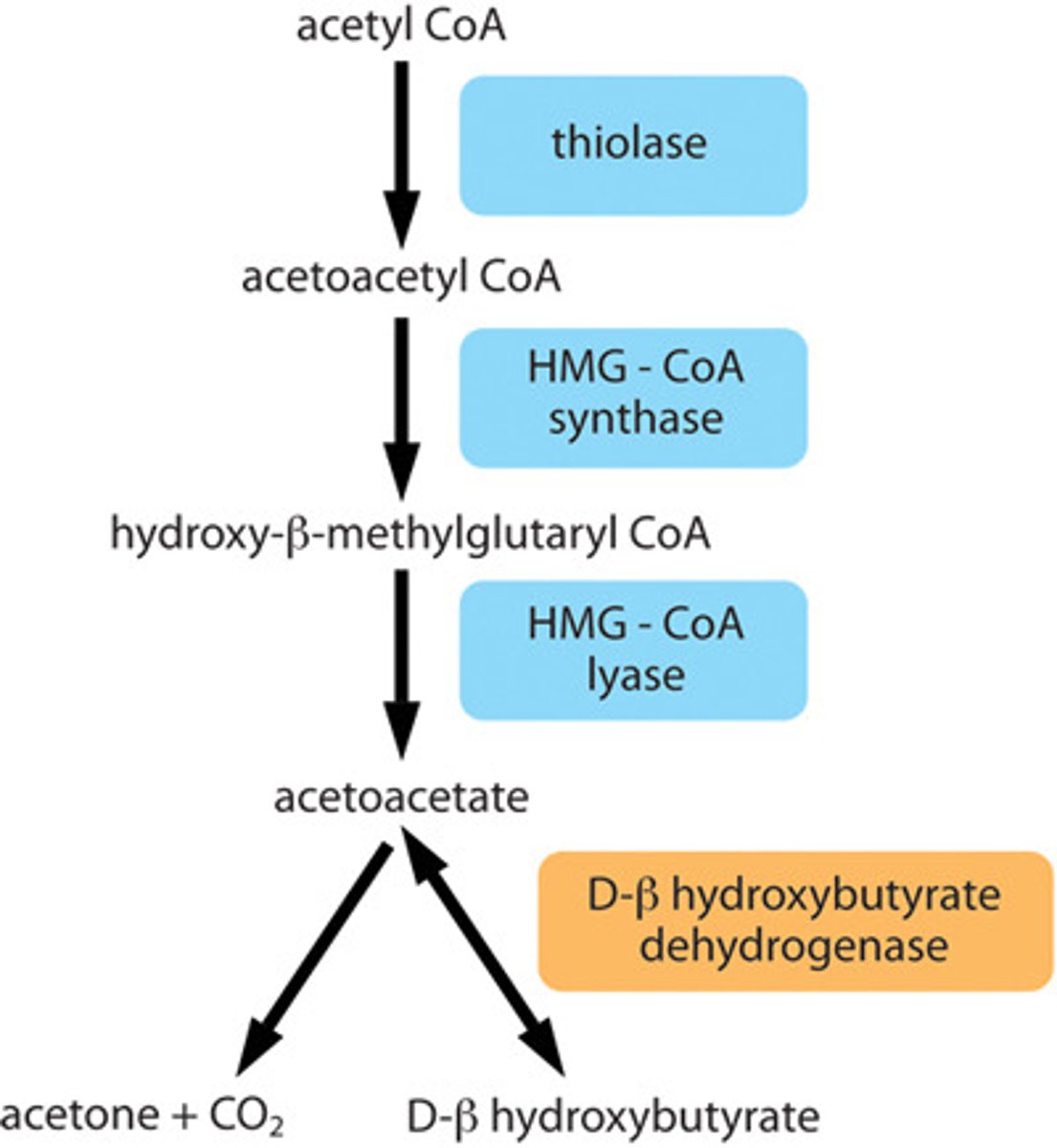
fatty acid ketogenesis
during starvation, glucose level fall and fatty acids are oxidized to supplement TCA cycle
in liver cells, remaining acetyl-CoA produced react together to form ketone bodies, enter brain or other organs to be reconverted to acetyl-CoA
2 acetyl-CoAs combined to form acetoacetate, which can split to beta-hydroxybutyrate and acetone
fatty acid synthesis
starts with acetyl-CoA and malonyl-CoA (from acetyl-CoA, using bicarbonate), activated to acetyl-ACP and malonyl-ACP
acetyl-ACP to acetyl-FAS (with fatty acid synthase attached)
fatty acid synthase helps combine malonyl-ACP with acetyl (release CO2), NADPH to remove ketone, then NADPH to remove double bond
in cytosol
need 2 NADPH for each 2 carbons added
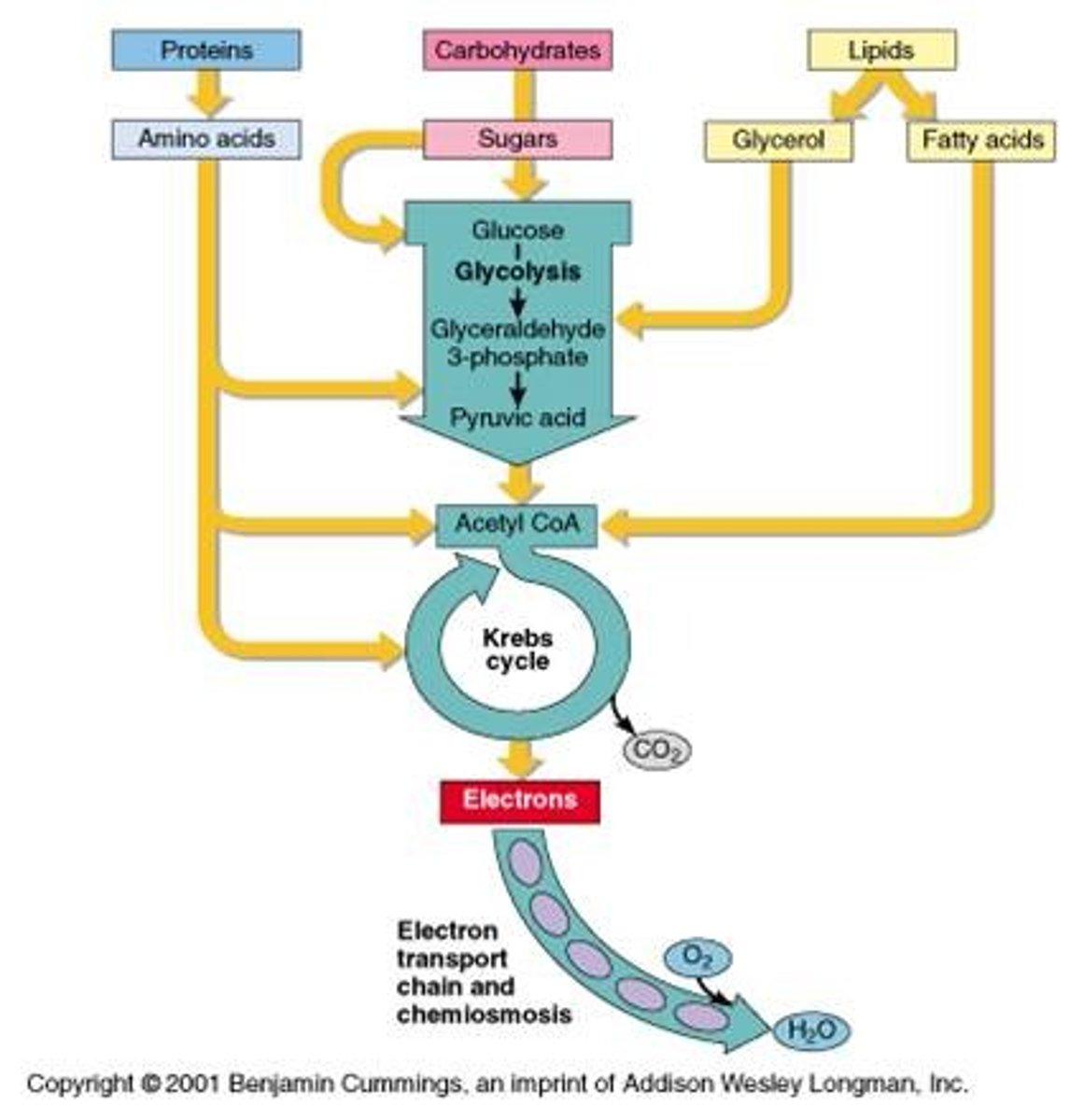
protein catabolism
protein broken down to amino acids by proteases
3 endpoints:
1. can be used to construct other proteins
2. amino end can be used for nucleotides or urea (excretion)
3. remaining carbon skeleton can be converted to acetyl-CoA or glucose
metabolic rate
how quickly an organism uses up stored energy reserves (protein, lipids, sugars)
metabolic states
absorptive state- glucose storage as glycogen in liver, fatty acid storage as triglycerides in adipose tissue, brain and muscle still using up glucose
post-absorptive state- glycogen broken down to glucose in liver, triglycerides broken down to acetyl-CoA to power TCA cycle and ketogenesis (which can enter brain)