BIMP10 del 2
1/92
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
93 Terms
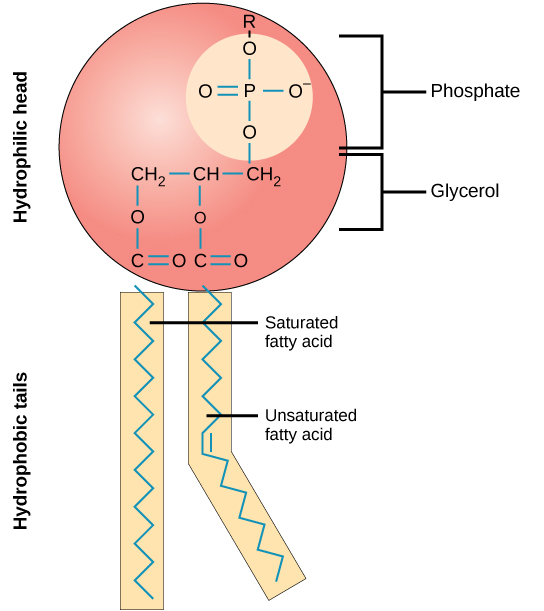
Phospholipid
2 fatty acids connected to a glycerol and a phosphate. Can be modified with choline.
Sphingomyelin
A lipid in the membrane that contains choline and is built from sphingosine. It is a phospholipid.
Glycolipid
Like sphingolipid but with a sugar group instead of the phosphor.
Charge of membrane
negative on inside and positive on outside. Phosphatidylcholine, one of the phospholipids, have an extra negative charge on inside.
Micelle
Shape that phospholipids naturally make to hide the hydrophobic parts and interact the hydrophilic parts with the water in the environment. Energetically favorable.
Unsaturated and saturated phospholipids
Unsaturated: double or triple bonds. Saturated: has no double or triple bonds.
Lateral diffusion
The phospholipids can move around in their layer easily.
Rotational diffusion
The pl can move around their axis.
Flip flop
when a pl moves to the other side of the membrane. It rarely happens because it requires energy. Need enzyme. Cholesterol does it easier than pl.
Lipid raft
Part of membrane that is more tightly packed than the rest. Usually has lot os sphingolipids and cholesterol. Can float freely in the membrane monolayer.
Glycocalyx
A coat of carbohydrates outside of the cell membrane, not all cells have it.
Epithelial cell
Common in skin and it lines the gut and kidney. has apical and lateral environments. Also has basal lamina.
Transport proteins: TP. Single pass proteins
a polypeptide chain that crosses the membrane as an alfa-helix
TP: multiple-pass proteins
polypeptide chain that crosses the membrane multiple times. is responsible for transporting water and other ions soluble in water.
TP: Transmembrane protein
goes through the membrane.
TP: membrane-associated proteins
only bound to the surface of the membrane by non-covalent interactions, alfa-helix or via lipid chain. some are peripheral membrane proteins and they can be detached by an extreme change in pH.
3 different modes of transport over membrane
passive diffusion, go from high concentration to lower.
facilitated diffuion (passive transport). will need help but can get through by channel or by transport protein.
active transport
Coupled transport
a type of active transport where 2 types of molecules are transported. symporter: both go in the same direction. antiporter: they go in opposite directions. the energy that is freed when one ion go with its gradient is used to push the other molecule. binding of either molecule increases the affinity to bind to the other.
ATP-driven pumps
Ex NA-K-pump or proton pump
Light or redix-driven pumps
active transport that bacteria use
Secondary and primary active transport
Secondary: ion-driven. the energy stored here is used to fuel the primary active transport. Ex in the NaK-pump: the first step when Na goes out of the cell. Primary: ATP-driven. Ex NaK-pump: the second step when K goes into the cell.
Explain why the cell cytoplasm has a pH and concentrations of different ions, that differ from the environment outside the cell
The transport and cell membrane makes it possible to keep two different environments stable aprt form each other.
Mitochondria
makes ATP. has its own membrane. it got its membrane from when it was an prokaryotic cell absorbed by an eukaryotic cell.
chloroplasts
makes ATP in plants
peroxisome
use enzymes to perform oxidative reactions. breaks doem fatty acids by this method.
cytosol
the fluid in cells. protein synthesis and intermediary metabolism is examples of what happens here.
nucleus
DNA and RNA. the fluid in the nucleus is not separated form the cytosol, has nucleoporins.
cytoplasm
the substance between the membrane and the nucleus (the core). the cytosol is the fluid part of the cytoplasm. the cytoplasm includes organelles and other parts, which float in teh cytosol.
KDEL
retention signal. a chain of aa that when attached to a protein inhibits it from leaving the ER, it is sent back there if it leaves.
transport cytosol→nucleus
small molecules can diffuse freely from the cytosol into the nucleus and macromolecules can enter nucleus by the nucleoporins.
nuclear envelope
the space between the nuclear membranes
nuclear basket
on the inside of the nuclear membrane. fibrils that form a basket that adapt to what is coming into the nucleus. the fibrils are on both sides but only on the nucleus side a basket is formed.
transport cytosol←→mitochondria
needed for enzymes to get into the mitochindria. the enzymes need to pass 2 membranes to get into mitochondria. protein translocator complexes recongnize signal sequences to direct a precursor to the right department. the proteins are unfolded when they are being transported. hsp70 maintain the proteins unfolded until they reach the right department and another set of hsp70 takes the proteins in. translocation is powered by ATP.
transport cytosol→peroxisomes
the proteins destined for the peroxisomes can go by the ER and get translocated by Sec61 or just come form cytosol. Ser-Lys-Leu (SKL) sequence on all proteins that go into peroxisomes. SKL is recognized by an import receptor in the cytosol. the protein goes through a protein translocator into the peroxisome, ATP-driven.
ER
proteins are folded. disulfide bonds are formed. N-linked glycolisation happens here. smooth ER synthesize cholesterol. it is also where proteins and lipids bud off and go to golgi or mitochondria. rough ER takes in proteins that are boing made.
cytosol→ER
ER-signal sequence directs proteins form cytosol to ER. For a ribosome that is translating an mRNA, the signal sequence gets cennected to an SRP and that bonds to a SRP-receptor in the rough membrane. the import is co-translational, the protein is not entirely done until the transport is over.
From the smooth ER where new proteins and lipids bud off and go to golgi or mitochondria.
Scramblase
can do a flip-flop for the pl in the membrane so the newly made pl can get to the lumenal leaflet( the membrane layer away from the cytosol)
Flippase
like scramblase bur for organelles. organelles can have different lipids on different sides of the membrane.
proteins from ER to golgi
new proteins ome from the ER and go on the secretory pathway to the golgi. they go in vesicles that are coeated with COPII. only rightly folded and assembled proteins leave the ER, otherwise they are sent back by chaperones. they come to cis-golgi where they are modified. they then leave by the trans-golgi by exocytosis.
golgi→lysosome
lysosomal hydrolases go to the lysosome. they hve M6P-groups that lead them to the lysosome. also have clathrin coats. M6P has a pH where it binds and pH where is releases, because of the acidic environment in the lysosome they are released there and can be recycled.
secretory proteins
for functions that need to secret much, fast. clathrin coated transport. they wait by the cell membrane for a signal ot be released from the cell, ex Ca2+ in nerve cells.
constituitive secretory pathway
go to the cell membrane from golgi. ex plasma membrane proteins. this is the default pathway, if no other signals are present the protein or lipid go this way.
regulated secretory pathway
for secretory proteins. they wait for a signal to be released ofrm the cell.
clathrin coat
from golgi, endosome and plasma membrane. consist of 3 heavy and 3 light chains and make a triskeleton. 36 triskeletons make a clathrin coat. they have adaptor proteins that select the cargo and bind to cargo receptors that bind to cargo.
COPII
coat that transport fom ER→golgi. GTPases help with assembly and disassembly when the vesicle reaches its goal.
COPI
coat that transport from golgi→ER. GTPases help with assembly and disassembly when the vesicle reaches its goal.
Rab protein
sit on vesicle and give fine specificity to determine where to land. is a GTPase. rab effector binds to rab protein on membrane.
SNARE protein
v-snare dock the vesicle by connecting to t-snare. catalyze fusion of membranes.
endocytosis
vesicles that come into the cell fuse with an early endosome. the early endosome has tubular projections. the late endosome does not have tubular projections.
endothelial cells
form lining of blood vessel. these cells are separated from the rest of the cells by a basal lamina. new blood vessels are formed by sprouting out form endothelial cells, process is called angiogenesis.
fibroblast
made from intermediate filaments. forms extracellular matrix and collagen.
actin filaments
can form microvilli in the intestines which increases food-uptake. also have a role in contracting muscles. has an ATP bound in the middle that it can use to build filaments. consist of 2 protofilaments wrapped around each other. is started off by Arp2/3 complex that is on cell membrane or on another actin filament. is split up by gelsolin, Ca2+ can activate gelsolin.
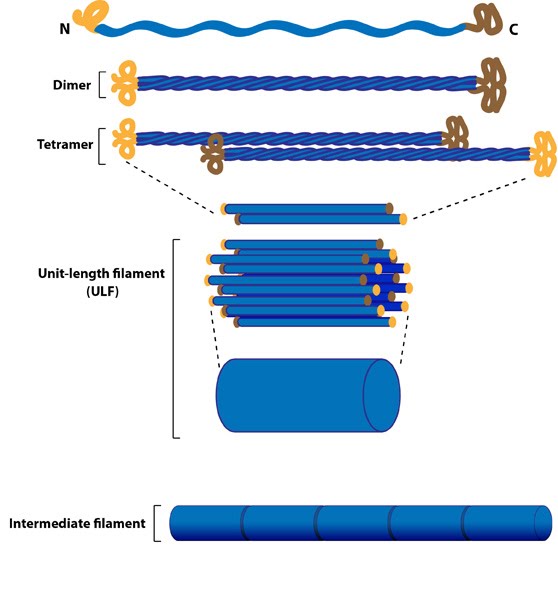
intermediate filaments
connects adhesive parts of cells to each other and to a basal lamina. can´t hydrolyse ATP but kinases can phosphorylate subunits which hydrolyse ATP. has an NH2 side and an COOH side. form fibroblasts.
microtubuli
global coordinate system. makes flagella. has + and - ends. varannan är alfa och varannan är beta. together they create a protofilament. 13 protofilaments make a microtubuli. each molecule can bind an GTP but only the beta ones can use it. is assembled with the help of Gamma-TuRC which is a gamma-ring complex that starts off the microtubuli. grows from - to +. MTOC: microtubuli organizing ring complex, can be a centrosome.
motor proteins
movement on cytoskeletal filaments (microtubuli or actin filaments) is mediated by motor proteins. they move through ATP. Ex myosin II, slides along actin filaments and makes muscles contract. myosin is activated by phosphorylation from MLCK.
kinesin
wanders on microtubuli. can move organelles.
flagella
Built from microtubules and dynein. Flagella is not a motor protein, moves by proton pump. makes sperm swim.
celia
Built from microtubules and dynein. like flagella but swims more whip-like.
prokaryotic cells
lack nuclear membrane, vesicular structures and mitochondria. have a cell wall. have ribosomes but different type from eukaryot cells. have single chromosome. can have flagellum. have plasmid: ring-shaped DNA with ca 5000 base pairs that float in the cytoplasm.
the cell wall is good for strength and it makes the bacteria have very different structures and shapes. it can keep out molecules.
prokaryotic cells show difference in protein modification, lacks some enzymes for ex glycolysation.
gram stain
classification of prokaryotic cells based off differences in cell wall structures. gram positive: thick layer of peptidoglycan (consist of amino acids and carbohydrates, makes a resistant cell wall). gram negative: have a thinner layer of peptidoglycan and have an outer membrane that gram positive does not have. have LPS outside of the outer membrane. periplasm separate the membranes and is also where the peptidoglycan is.
protein set for degradation
proteins are marked for degradation by attachment of ubiquitin to a Lys residue. more ubiquitins are then added on and they are recognized by protease complex, called proteasome.
nucleoside
a nitrogenous base bound to a sugar.
nucleotide
a nitrogenous base, a sugar and a phosphate
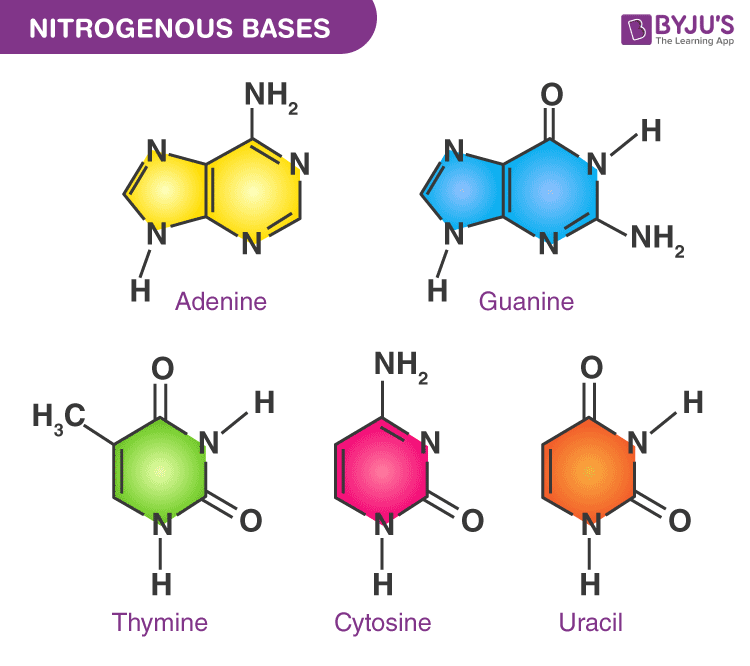
nitrogen bases
ska kunna rita ut alla olika
nucleolus
where ribosomes are made. is a part of nucleus.
chromatin
DNA bound to histones and other proteins. The DNA is negatively charged and the histones are positevly charged.
nucleosomes
DNA wrapped aound 8 histones.
centrosome
organizes microtubuli
heterochromatin
areas that are extra dense in DNA in chromosomes
mitochondrial DNA
17 000 base pairs
purine and pyrimidine
purine is 2 rings and pyrimidine is 1 ring
types of mutation in DNA
base substitution
deletion
insertion
frameshift: one of the strands gets nucleotides added or deleted in numbers that are not multiples of 3
centromere
center of chromosome. there is also a p band and a q band.
reciprocal chromosomal translocation
abnormal recombination can lead to 2 double helices crossing over so chromosomes trade DNA
mouse chromosomes
20 pairs, 19 regular + x and y
human chromosomes how many
23 pairs
telomere
end of chromosome. allows chromosome to be efficiently replicated. it is usually 7 nucleotides repeated about 1000 times, for humans GGTTA. an enzyme called telomerase makes it. telomerase has reverse transcriptase which is an enzyme that uses an RNA template to make an DNA strand. telomerase synthesizes the lead strand 3´→5´ and then the lagging strand is synthesized by DNA polymerase.
replication origin
where replication starts in a chromosome. chromosomes have multiple replication origins.
kinetochore
protein complex that forms at the centromere after duplication and attached the chromosomes to the mitotic spindle and then the sister chromatids are pulled apart.
histone
consist of two dimers, H3-H4 and H2A-H2B connecting. have 8 terminals that protrude from it and help hold the nucleosomes together. during replication the histone is broked down into H3-H4 and H2A-H2B pairs, the first stays loosely bound to the DNA replication fork and the second disappears away and new ones replace them.
PCR
Technique used to make multiple copies of DNA segment of interest from a small initial sample. Makes detection of pathogenic virus or bacteria, identification of individuals possible.
Put primer, polymerase, nucleoside triphosphates and DNA in an PCR machine. also need suitable buffer and Mg2+ as a cofactor. DMSO is a good solver, polar aprotic. at 95 degress the DNA gets denatured. then at 55 degrees (annealing) the primers bind to the DNA where they fit. the primer is 5´→3´ and binds to the DNA 3´→5´ then at 72 degrees the polymerases binds the primers and starts making copies with the free nucleosides.
lagging strand
DNA polymerase can only work in ne direction, 5´→3´. the lagging strand is the opposite, 3´→5´. it is synthesized in the opposite direction from primers that attach on multiple sites on the DNA. the fragments created are called okazaki fragments. they are put together by DNA ligase. DNA ligase uses ATP to bind the 5´ phosphate group to the 3´ OH-group.
topoisomerase
enzyme that helps unwind the DNA and relaxes it when it is being decoiled. used during replication and transcription.
proofreading of DNA (replication)
base-pairing: before nucleotide is added to new strand DNA polymerase tghtens the active site. if the active site is not tight enough it could be the wrong nucleotide and it diffuses.
exonucleolytic proofreading: DNA polymerase needs the 3´ end of the nucleotide to continue. if the wrong is added- if 3´→5´ growth would happen- DNA polymerase can´t extend that strand because there is no cleavage of phosphate groups and therefore no enrgy is released. a proofreading exonuclease works 3´→5´. when an incorrect base pair is recognized, DNA polymerase reverses its direction by one base pair of DNA and excises the mismatched base. then the polymerase can re-insert the correct base and replication can continue.
MutS and MutL scans the DNA for errors in newly made strands. they bind at the error and can easily break the strand there because the error bonds weakly. then DNA polyermase fixes it and DNA ligase seals the strand.
DNA primase
RNA primers are made from DNA primase, which is an RNA polymerase, which uses DNA strands as template. the RNA primers are then used to crete okazaki fragments. they are ca 10 nucleotides long and are attahced every 100-200 nucleotide on the DNA.
DNA helicase
single-strand DNA binding proteins. they help open up the DNA to make a replication fork. uses ATP to move along DNA and break hydrogen bonds. is a hexamere (has 6 subunits).
replication fork
protein complex that DNA gpes through when being replicated.
sliding clamp
circles arounf the DNA polymerase and gives high processivity.
initiator proteins
start the replication by the replication origins.
depurination
when the bond to the sugar breaks by hydrolysis. happens to purines. can lead to loss of a base pair or that replication mahcinery chooses another, wrong, nucleotide. the daughter cell will also get this.
deamination
spontaneous cytosine→uracil. the NH2 gets replaced by an O through hydrolysis. can lead to substititution, uracil binds to adenosine. daughter cell also gets this.
UV damage to DNA
If the cell is exposed to Uv light: adjacent bases can form dimers, hydrogen bonds. If not corrected- can lead to deletion of base pair or base pair substituted in daughter cell when replicated.
repair of spontaneous DNA damage
base-excision repair: DNA glycosylases are specifie in a type of damage. find the errors by traveling along the DNA.
nucleotide excision repair: repairs large changes, ex the UV damage. can repair many different types of errors. scans helix and cleaves,, then DNA polymerase repairs and DNA ligase seals.