Lecture 9- DNA repair mechanisms
1/26
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
27 Terms
What repair mechanism is used for alkylation?
Direct reversal
What repair mechanism is used for deamination and oxidative damage?
BER
What 2 repair mechanism is used for UV light?
Direct reversal, NER
What repair mechanism is used for replication errors?
MMR
What repair mechanism is used for x-rays?
non-homologous end joining.
What is the roll of P53 tumour suppressor and how does it carry this out?
Responds to/detects DNA damage. If damage is detected, it arrests the cell cycle at the end of G1, allowing for time to repair. It also activates DNA repair proteins.
What does P53 trigger if damage is too severe?
Apoptosis or senescence.
DIRECT REVERSAL/REPAIR
What type of damage does this deal with? Give an example.
When the damage has converted A,C,G,T to something else, it may be possible to convert back to the original nt.
Example: Demethylation after alkylation.
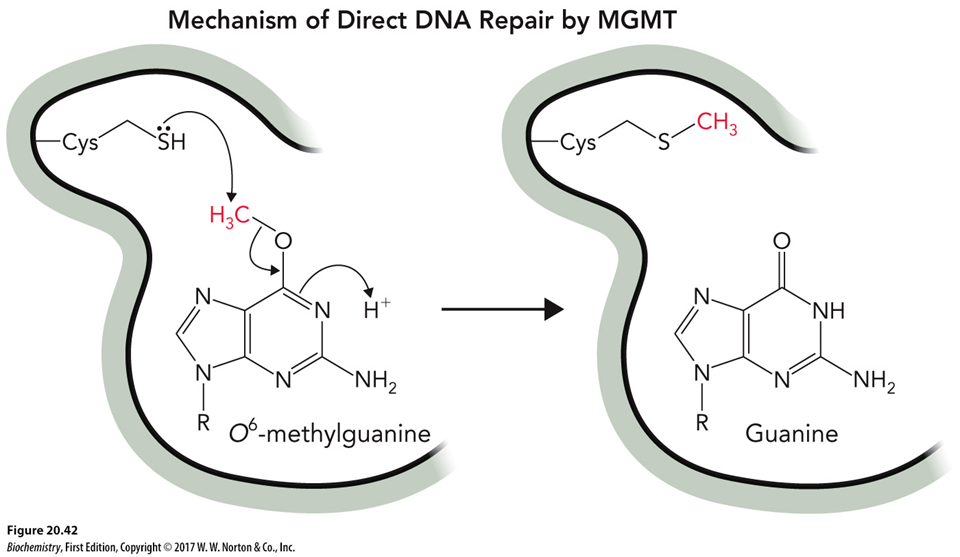
How does demethylation fix damaged DNA?
Specific reversal proteins, e.g., O6-methylguanine-DNA methyltransferase (MGMT). This transfers methyl/ethyl group from G to a Cys residue on itself so G is restored.
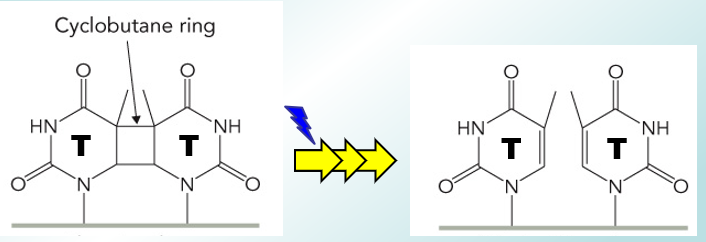
How does photolysis of dimers (photoreactivation) repair damaged DNA?
It removes crosslinks following UV light damage. DNA photylase is used to absorb blue light and break T-T internucleotide bonds, using FADH → 2 Ts restored.
This isn’t used in mammals as they don’t have this enzyme.
BASE EXCISION REPAIR
What is BER? How does this occur?
This removes individual bases (local correction).
A group of more than 6 DNA glycosylases recognise abnormal bases and cleave them from deoxyribose, creating an abasic site.
What is the role of glycosylases during BER?
They flip out bases for closer checking. If the base is found to be incorrect, the sugar-base bond is cleaved, but UDGase remains attached to DNA.
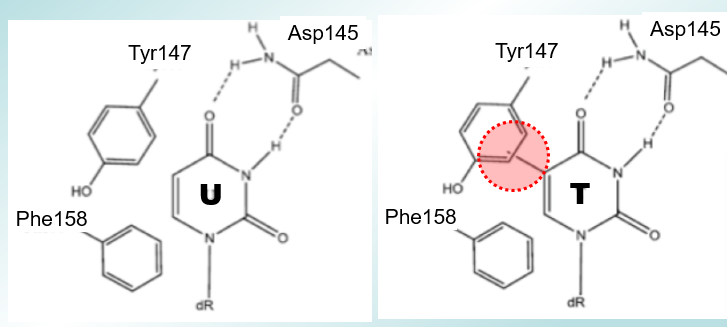
How does UDGase discriminate between U and T?
There is a steric clash of methyl group on T with Tyr residue. There is a seperate TDGase for GT pairs. If Tyr147 is mutated to Ala, both T and U will be removed by the enzyme.
What are the steps used in BER to repair DNA?
U in DNA
Base removed by Uracil-N-glycosylase
Baseless nt recognised and phosphodiester backbone cleaves by AP endonuclease
Nicked DNA
Pol I nick translation restores T + DNA ligase seals nick
Pol B in eukaryotes instead of Pol I.
NUCLEOTIDE EXCISION REPAIR
How does NER repair damaged DNA?
Oligonucleotide fragments are removed from 1 strand (bulk correction), triggered by changes in duplex structure due to damage. Corrects any type of sequence damage.
What protein complex is used in NER?
UvrABC exinuclease in E.coli
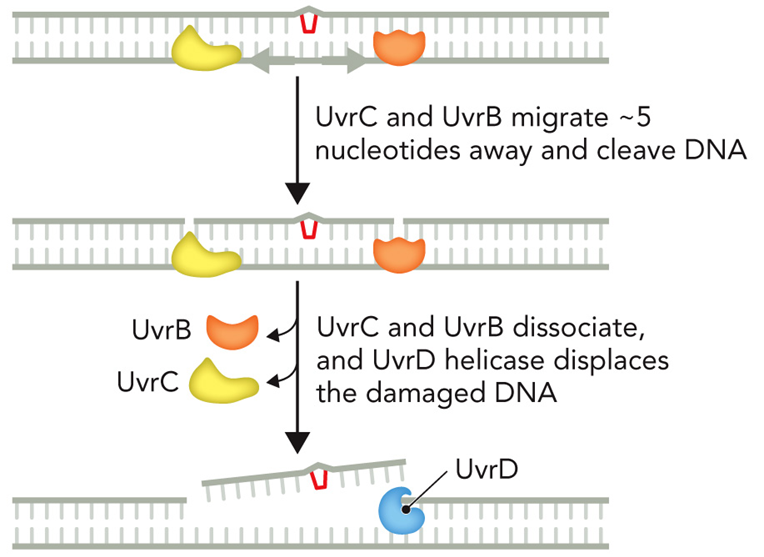
What is the mechanism for NER?
Thymine dimer in DNA sequence
UvrA and UvrB bind to thymine dimer
UvrA subunits dissociate and UvrC subunit binds.
UvrC and UvrB migrate 5 nucleotides away and cleave DNA
UvrC and UvrB dissociate, and UvrD helicase displaces the damaged DNA
METHYL- DIRECTED MISMATCH REPAIR (E.COLI)
How does this repair damaged DNA?
Detects and removes incorrect base pairs.
How does this take place?
a mismatched pair will distort the helix
this can be detected by specialist proteins
incorrect base removed
To find out which one of the 2 is wrong, must use strand directed mismatch repair.
Role of hemi-methylation?
Provides information on which strand is parent (correct) and daughter.
What is the mechanism for MMR?
MutH binds to unmethylated GATC at OriC, identifying the daughter strand.
MutS binds to a distorted site on the duplex.
MutL binds to MutS.
MutL/MutS complex travels back to the origin and activates MutH.
MutH cleaves daughter strand (nicked).
Specialised helicase (UvrD) and exonucleases remove nt until past the distortion.
Pol III fills in missing nt. DNA ligase seals nick.
For MMR, what don’t eukaryotes have?
No homologues of MutH, but they don’t use hemimethylation tags either.
8-OXO-G
Role of MutT?
Recognises 8-oxo-GTP and hydrolyses it.
Role of MutM
recognises 8-oxo-G in DNA and removes it via BER
Role of MutY?
Recogises 8-oxo-G opposite A in DNA and removes the A, via BER
DISEASE
What is Xeroderma pigmentosum (XP)? What causes it?
Individuals show dry, parchment-like skin and many freckles.
Increased sensetivity to UV light
1000-fold increased risk of skin cancer.
Causes: Inherited defect in 1 of 8 distinct genes responsible for components of the NER complex.
What is Hereditary non-polyposis colon cancer? How does it occur?
Individuals exhibit a predisposition to colon cancer. Leads to accumulation of mutations throughout genome.
Cause: Defect in human equivalent of MutS/L MMR system (MSH2 and MLH1).