lf206 lecture 6 - eukaryotic genomes, post transcriptional control
1/31
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
32 Terms
name 3 eukaryotic polymerase II post-transcriptional events
why do these events not always occur?
5’ capping
splicing
3’ end processing
they require factors that bind to the phosphorylated C-terminal domain of RNA Pol II
what does CTD stand for?
it is found on __karyotic RNA Pol __
it is comprised of a 7 amino acid sequence → what is it?
phosphorylation of ____ 2 and 5 marks the transition from transcriptional initiation to _______
C-terminal domain
found on Pol II
YSPTSPS
seryls
elongation
what is transcriptional initiation and elongation?
initiation → first steps of transcription where RNA polymerase unwinds DNA and begins to read to synthesise RNA
elongation → RNA polymerase building RNA in 5’ to 3’ direction
addition of 5’ cap is an early event (nascent mRNA ~ __bp)
it distinguishes Pol II ______ from other RNA molecules
stabilises RNA, there is no 5’ _____, so it is resistant to 5’ _________
aids in further processing and export to the ________
is required for efficient ________ of mRNAs
25
transcripts
phosphate
exonucleases
cytosol
translation
the 5’ cap is a modified ________ nucleotide
name the 3 capping enzymes - these enzymes are recruited by _____
guanine
RNA triphosphatase
guanylyl transferase
methyltransferases
the phosphorylated C-terminal domain (CTD) of Pol II
a multi-subunit ________ ATP-________ decapping enzyme ______ removes the 5’ cap
it restores the 5’ ______ on the mRNA
now the mRNA can no longer be bound by ______ so cannot be translated
the mRNA is ______ by a 5’-3’ RNAse
cytosolic
dependent
complex
phosphate
ribosomes
degraded
what is rDNA?
ribosomal DNA that encodes for rRNA (small subunits that make up a ribosome)
Tetrahymena thermophilia rDNA encodes for two large ribosomal subunits (___ and ____) and a small ribosomal rRNA (___)
28S 17S
5.8S
what is splicing? how does this relate to group I introns?
removing introns from pre-mRNA transcript to only have exons making mature mRNA
group I introns can self-splice
Cech discovered that an ______ in Tetrahymena thermophilia rRNA could remove itself - this is called?
the steps:
the intron ____
a _______ co-factor (GMP, GDP, GTP) is held in a pocket
the 3’ -__ of the co-factor is a nucleophile that attacks the ______ at the 5’ splice site
the 3’-OH of the upstream ____ attacks the phosphate of the 3’ _____ site
the ______ are fused and the intron in degraded.
introns
self-splicing
folds
guanosine
OH
phosphate
exon
splice
exons
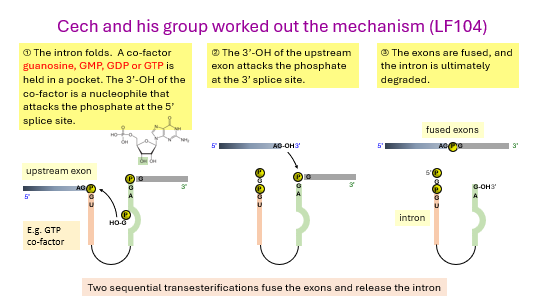
group I introns are ___-splicing
group II introns utilise 2’-__ group of the branch site ______ as an in-built co-______
both group I and II introns encode protein maturases - what are these?
nuclear spliceosomal introns (what are these?) use the same catalytic mechanism as group __ introns.
self
OH
adenine
factors
proteins that increase efficiency of excision by stabilising folded 3D structure
introns found in pre-mRNA of eukaryotic cells to be removed by the spliceosome
II
how are eukaryotic introns spliced?
introns ___ and the 2’-OH of the _____ ___ adenosine attacks the phosphate at the _’ splice site
adenosine now has 3 _____________ bonds. one includes a strange bonds which is?
the 3’ -OH group of the _____ exon attack the _______ at the 3’ splice site
the exons are ____ and the intron is released as a ______
fold
branch site
5
phosphodiester
2’,5’ phosphodiester bond
upstream
phosphate
fused
lariat
the spliceosome is an RNA and _____ complex
it is composed of 5 subunits called ______. name all 5
what 3 specific sequences does the spliceosome recognise?
protein
snRNPs (small nuclear ribonucleoproteins)
U1, U2, U4, U5, U6
5’ splice site, branch site, polypyrimidine tract (poly Y)
where are the following located?
5’ splice site, 3’ splice site & branch site
5’ splice site: beginning of intron
3’ splice site: ending of intron
branch site: located upstream from ending (3’ splice site) of intron
spliceosome mechanism
U1 snRNP binds to the _’ splice ___
_____ _____ _______ (BBP) and the protein U2 auxiliary _____ (U2AF) bind the branch site and associated pyrimidine tract.
a trimer of U4, U5 and ___ snRNPs associates with the complex
U2 snRNP is recruited and displaces ___
base pairing of U2 snRNP with the ______ causes the branch site _____ to bulge towards the _’ splice site
loss of _ and _ snRNPs activates the complex
the 2’ -__ of the branch site adenine attacks the 5’ ____ site phosphate
the ____ is formed
U6 snRNP guides the 3’-OH of the upstream ____ that attacks the 3’ _____ site phosphate
5, site
branch binding protein, factor
U6
BBP
intron, adenine, 5’
OH, splice
lariat
exon
splice
how does U1 snRNP find the 5’ splice site?
U1 is already close to the RNA Poll II tail - what is it called?
capping of the pre-mRNA transcript by the _______ enzyme (CE) is an early event
U1 snRNP is recruited by the _________ CTD
U1 snRNP _____ the growing mRNA for the 5’ ____ site and binds it
C-terminal domain (CTD)
capping
phosphorylated
scans, splice
what does U2AF do.
it is bound by the ___ of RNA Pol __
whilst the pre-mRNA chain is growing, BBP/U2AF scans for __________ and _________
once recognised, the ________ is assembled
true or false - it is apart of the spliceosome structure?
CTD, II
pyrimidine tract and branch site
spliceosome
false
splicing by spliceosome is regulated by _______ sequences.
individual motifs (what are these?) have different affinities for spliceosome components.
consensus
motifs - recurring patterns (can be nucleotides, can be amino acids)
biogenesis of U1, U2, U4 and U5 snRNPs
transcription and _______ in the nucleus
_______ loading of a ring of ____ Sm proteins onto the snRNA by ___ proteins
the cap is __________
transport to the _____ and maturation by association with other proteins
U6 lacks __ proteins so remains nuclear.
capping
cytosolic
7, SMN (Survival Motor Neuron)
hypermethylated
nucleus
Sm
function of the protein components of snRNPs
stability of the snRNP
difference between snRNA and snRNP
snRNA: just a small sequence of RNA (e.g. U1, U2 etc.)
snRNP: snRNA + associated proteins that can then catalyse intron removal (e.g. U1 snRNP, U2 snRNP etc.)
exons are defined by the binding of proteins during ________
this allows U1 to find __________
SR protein (rich is Ser and ___) bind to _____ ______ ______ (ESEs)
splicing ______ flank the 5’ and 3’ ____ sites ensuring precise intron removal
________ hnRNP proteins can bind to exon sequences and act as _______ repressors as they compete with __ proteins.
transcription
5’ splice site
Arg, exonic splicing enhancers
factors, splice
heterogenous nuclear ribonucleoproteins, splicing
SR
what is the poly Y tract involved in?
alternative splicing
what is alternative splicing & what is it also called?
a single gene can produce multiple different mRNA transcripts
exon skipping
a strong consensus poly Y tract leads to?
strong binding of BBP and ____
BBP is displaced by __
results in a _’ end of an intron strongly defined → normal splicing
U2AF
U2
3
a weak consensus poly Y tract leads to?
BBP and ____ binds
U2 is _____
3’ end (which part of the intron?) of intron defined and you get normal _____
OR
U2AF ______
BBP and __ are not bound
_’ end of intron is not defined
_________ scans for the next U2AF and exon is _____ (alternative splicing)
U2AF
recruited
the beginning of the intron, splicing
disengages
U2
3
spliceosome
alternative splicing is regulated by the sequence of _______
poly Y tract (polypyrimidine tract)
give an example where splicing goes wrong
Alexei (great grandson of Queen Victoria) had a G residue in blood clotting factor IX
no masking the mutation due to having only 1 X chromosome
this caused splicing to occur 2 nucleotides early and produced an ineffective blood clotting factor
Anastasia had a heterozygous A/G genotype in blood clotting factor IX
therefor has a normal phenotype as the disease is recessive
how is the poly(A) tail added?
GU rich region is cleaved leaving 3’ -OH
the GU rich region is degraded in the nucleus
roughly 250 adenine residues are added at the 3’ end
how is the GU rich region cleaved?
RNA is folded to bring _ key sites together
the poly(A) tail is bound by __ ____ __ ___ (CPSF)
the cleavage site is bound by _______ _______ (CF)
the GU rich region is bound by ____ ____ ____ __ (CStF)
these provide a recruitment site for the polymerase called ___
cleavage provides a 3’-__ for PAP which adds ______ residues
this is a very ____ process
3
cleavage and polyadenylation specificity factor
cleavage factor
cleavage stimulation factor F
PAP
-OH, adenine
slow
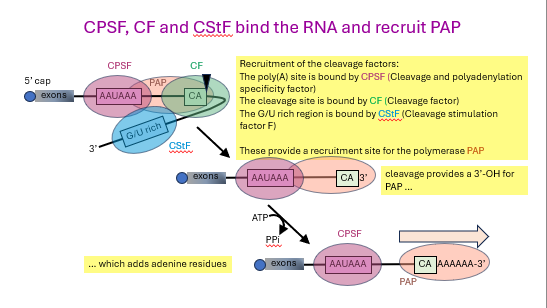
how to go from slow adenylation to fast adenylation? [3]
PAP slowly adds nucleotides
poly(A) binding protein then associates and extends tail faster
~200 nucleotides are added and PAP disengages
functions of polyadenylation
nuclear transport → nuclear export receptor is loaded onto mRNA by Pol II CTD (a ‘ticket’ to leave the nucleus)
translation → pseudo circularisation (making loop) of transcript promotes translation (poly(A) tail & PABP interact w/ methylated 5’ cap)
stability → mRNA often degraded from 3’ end first, tail extends lifespan by reducing degradation rate