BM210 - BLOCK B (not L1)
1/166
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
167 Terms
how do catalysts increase affect reaction? what is the transition state?
increases rate of reaction by lowerig activation energy (transition state)
is not consumed in the reaction
does not effect equilibrium
transition state is the unstable point that reactants must pass through to become products. → catalysts stabilises this by lowering the activation energy it occurs at
how is the active site formed?
Folding of the protein brings side-chains of various amino acids far apart in primary squence into close juxtaposition, forming an active site.
what is the process of an enzyme catlysed reaction?
substrates enter active site (Enz + S)
substrates are held by ionic interactions and hydrogen bonds → Enz-S complex
enzymes breaks/forms bonds in substrates converting to products → Enz-P complex
products are released (Enz + P)
ezymes remained unchaged and reaction can start again
reversible reaction
Enz + S ⇌ Enz-S ⇌ Enz-P ⇌ Enz + P
how does the active site lower activation energy/stabilise transition state?
positions substrates in correct orientation/alignment → avoiding collison
complementary to transition state (not substrate necessarily) → stabilising the transition state
amino acid side chains of active site stabilise electron distribution through hydrogen bonding, ionic interaction, covalent interaction → stabilising transition state
substrate is strained (distorted) → stretching/bending pushed closer to transition state faster
what are the Non-covalent interactions between the substrate and the amino acid side-chains of the active site?
Øacidic groups (Asp, Glu) → ionic bonds
Ø basic groups (Lys, His, Arg) → ionic bonds
Ø hydrophilic interactions with –OH or (Ser, Thr, Tyr)
Ø hydrophilic interactions with –SH (thiol) or (Cys)
Ø hydrophilic interactions with amide groups (Asn, Gln)
Ø aromatic interactions (Phe, Tyr, Trp)
Ø hydrophobic interactions (Ala, Leu, Ile, Val, Met)
how do reactive groups at the active site catalyse?
Ødonating (break bonds) or withdrawing electrons (form bonds)- from amino acids
Ø stabilising or generating free radical intermediates - weak interactions and temporary moving of electrons
Ø forming temporary covalent bonds → (a transition state intermediate) - acyl-enzyme → lowers activation energy
example of a enzyme-substrate interation?
yeast henokinase from hexokinase (enzyme) + glucose (substrate)
what is the induced fit model?
the enzyme changes shape around the substrate - the enzyme is flexible not rigid
what are co factors?the 3 types and their involvement with enzymes?
co-factors are non-proteins molecules
metal ions (inorganic) - Mg²⁺, Zn²⁺, Fe²⁺, Cu²⁺ → stabilise negative charges
prosthetic groups (organic) - attached to enzyme covalent bonds → heme, lipoic acid
co-enzymes (organic) - binds loosely to enzymes - NAD+/H, FAD, coenzyme A → carry chemical groups or electrons
what is the difference between a holoenzyme and apoenzyme?
holo - enzyme + cofactors → catalytically active
apo - enzyme with no cofactors → cataclytically inactive
what are the 6 major classes of enzymes and action?
1. Oxidoreductases - redox - dehydrogenase
2. Transferases: transfer a chemical group from one substrate to another - kinase
3) Hydrolases: hydrolysis (water splits the bond) of C-O, C-N, O-P and C-S bonds - phosphatases
4) Lyases: addition across a carbon-carbon double bond
5) Isomerases: intramolecular rearrangements
6) Synthetases: formation of bonds between two substrates
frequently linked to utilisation of ATP
unit of enzyme activity
(EU) = 1 μmol min-1
specific activity
acitivty per mg of protein - gives protein purity (μmol min-1mg-1)
hyperbolic reaction rate
rate of product formed slows - - from denaturation, substrate depletion
isoelectric point (pI)?
is the pH at which the molecule has no net electrical charge.
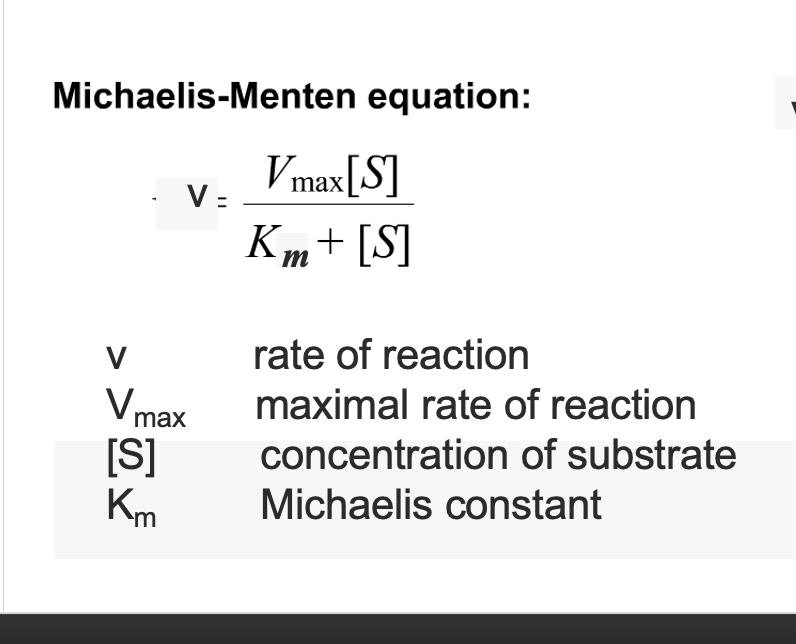
michaelis constant
Concentration of substrate to achieve half the maximum
rate of the reaction is Km
Low Km → high affinity
(only a small amount of substrate is needed to reach half Vmax)High Km → low affinity
(requires more substrate to reach half Vmax)
Michaelis-Menten equation
ternary complex? (sequential mechanism vs ping pong)
enzyme holds both substrates simultaneously
sequential - Both substrates must be present at the same time at different active sites - increasing B increases afinity for A (substrates are dependent) → converging lines on Lineweaver–Burk plot:
ping pong (double displacement) - substrates bind one at a time - substrate becomes modified following binding of 1st substrate ( substrates are independent) → parallel lines on Lineweaver–Burk plot:
Lineweaver–Burk plot:
double reciprocal plot used to determine Km and Vmax - straigns hyperbolic mentalis menten curve to be more accurate
Y-intercept = 1 / Vmax → vmax increases moving up
X-intercept = –1 / Km → km increases moving right
allosteric enzymes
produces sigmoidal curve - cooperative binding
multiple subunits with identical active sites
eg cyclin dependent protein kinase
example of covalent modification of an enzyme?
phosphorylation of ERK2 at threonine and tyrosine results in a structural change of the activation loop exposing hydrophobic region
reversible vs irreversible inhibitors?
Øreversible inhibitors - non-covalent binding to ezyme, unspecific → blocks substrate binding
Øirreversible inhibitors (inactivators) - bind to enzyme covalently, are substrate analogues, part of reaction → transition state covalent intermediate does not break down
competitive inhibotr? (reversible?)
competes with the substrate for binding at the active site -
REVERSIBLE - as they bind non covalently and siplaced with high conc of substrates
km increases because substrate ability to bind decreases
mixed inhibitors
bind allosteric site of E or ES complex chanigng enzyme shape → loweing vmax and altering km
can be competitive binding straight to E → km increases vmax decreases
can be UNcompetitive binding ES → km decreases VMAX DECREASES
can be NONcompetititive - binds E and ES - unchanged km / vmax decreases
allosteric activator/ inhibitors ?
sigmoidal - dont follow michael mentalis
not km → k0.5 = substrate conc at ½ vmax
positive modulator/activator → lower k0.5
no modulator - k0.5 unchanged
negative modulator/activator → increased k0.5 - more substrate need to reach ½ vmax
aldehyde vs ketone
aldehyde - HC=O - end of a chain
ketone - C=O - in the middle of a chainhem
what makes ribose deoxyribose
missing OH on carbon 2
hemiacetyl formation ? in glucose - pyronose?
aldehyde group on C1 reacts with alcohol OH group on C5 joining to form a/b anomers ring form
a = OH below
b = OH above
anomeric carbon is c1
hemiketal?
hemiacetyl formation but a ketone group not aldehyde
seen in fructose - ketose
anomeric carbon is c2
Sorbitol
Formed by the reduction of the aldehyde group of glucose to a hydroxyl group.
→ CHO → CH2OH
sorbitol is a sugar alcohol - tastes very sweet
o vs n glysosylic bond - in dna?
O bond formed when anomeric carbon reacts with alcohol eg methanol or serine
→ C1 with H above and OCH3 below
N bond formed when anomeric carbon binds nitrogenous base or lysine
→ C1 with H above and NR2 below
in dna - phosbate bind with O-glycosylic bond on C5 and N-glycosylic bond on C3
are phosphorylated sugar negative? examples
yes → cant pass cell membrane without transporters
G6P - first step of glycolysis
DHAP - metabolism
GAP - oxidised in glycolis to make ATP
which sugar type is nutritionally important\?
hexoses - glucose , fructose, galactose
disaccharide formation
O-glycosilic bond between two monosacharrides
intrinsic sugars vs extrinsic
intrinsic - good sugars contained within plant cell walls
extrinsic - bad, free in solution eg plaque - provide food for bacteria
— lactose from milk is good though
3 most common disacharrides and their bond type plus isotype of maltose
sucrose (cane or beet sugar - made from one glucose and
one fructose) - a 1-2 link
Trehalose – especially in mushrooms - a 1-1 link
maltose (made from two glucoses) - produced by germinating cereals eg barley via amalase - a 1-4 link
→ isotype is isomaltose — linkages at a1-6
what moleculaes make up starch? glycogen similarity?
Amylose – chain of glucose molecules (a-1,4)
Amylopectin – chain of glucose molecules (a-1,4), every 30th glucose → branch to other glucose residues (a-1,6 — ISOMALTOSE!)
glycogen - similar to starch, but branch every 10th glucose via a-1-6
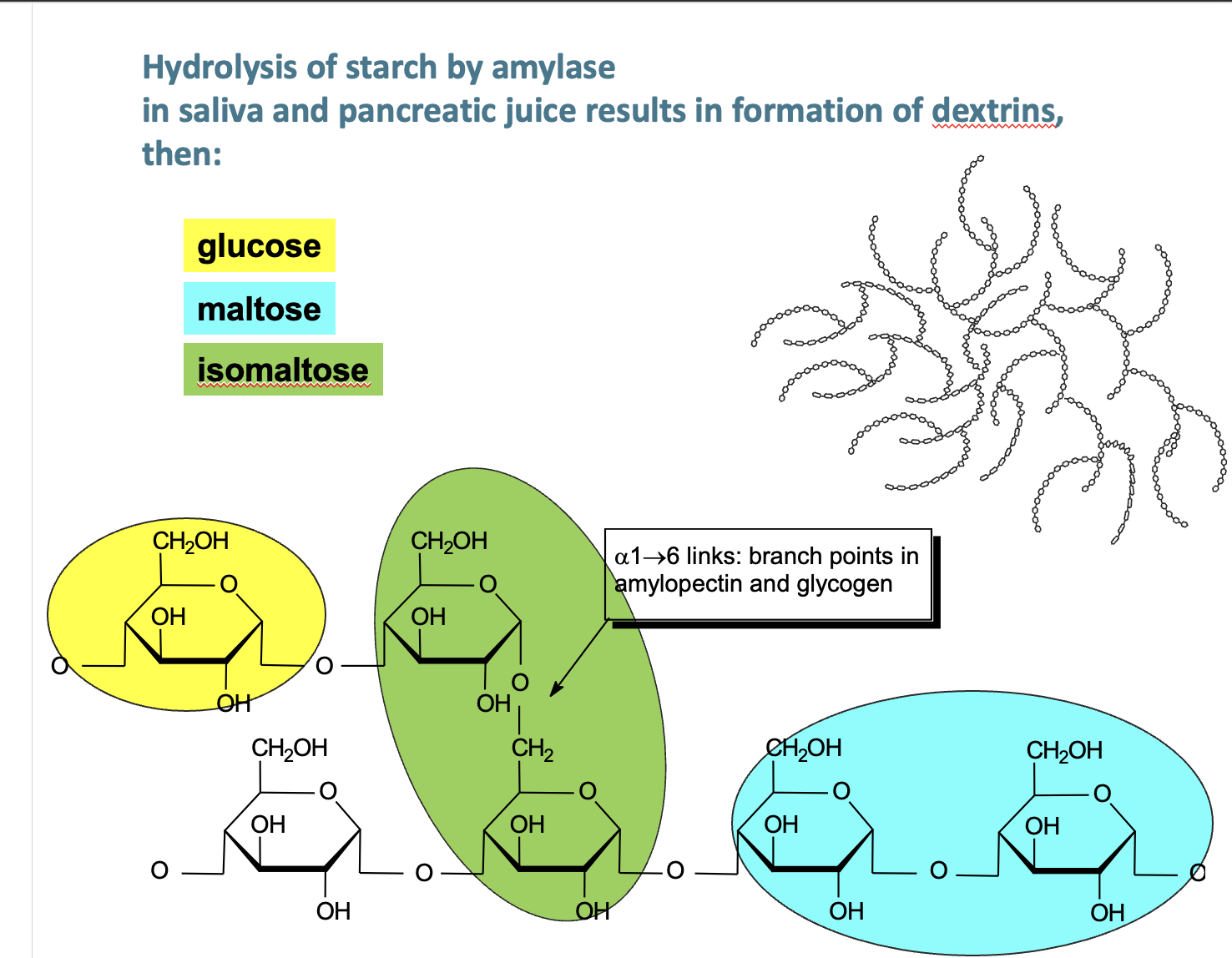
non - starch polysachharides?
e.g. Cellulose (glucose linked b-1,4
lactose intolerance
lactaste hydrolyes lactose at b1-4 link into glucose and galactose
→ decreases in lactase activity with age means lactose is converted to lactate, methane and hydrogen gas → farts (methan and hydrogen) and diarrhoea (osmosis from lactate)
glycogen metabolism
storage form of glucose in liver mostly (feeds brain) and skeletal muscle → insulin = store glucose, glucagon = release glucose
reducing / non-reducing ends of polysaccharides
reducing - free anomeric carbon can donate electrons
non reducing - anomeric carbon involved in glycosyllic bond so cant reduce
what is more stable open chain or hemiacetyl?
hemiacetyl
acetyl def
molecule with two single bonded oxygens attached to the same carbon atom - ie c1 with O-glycosillic linkage seen in lactose (b1-4) → is the non reducing end
why doesnt glycogen have a reducing end?
final glucose residue is covalently bound to a protein termed glycogenin via tyrosin
what is glycogenin
glycosyltransferase dimer at core of glycogen
what enzymes does glycogen contain
glycogen synthesis (glycogenesis) and degradation (glycogenolysis).
→ act on non reducing ends n
post translational modifications of carbohydrates
glycolysation - helps glyco protein stability, folding, recognition, nutrient sensing
protein O-glycosylation at serine or threonine in the Golgi
protein N-glycosylation at asparagine (Asn-X-Thr/Ser motif) in the ER sugar added via n-acetylglucosamine (GLcNAc) THESE ARE N LINKED GLYCANS
proteoglycans
proteins with long glycosaminoglycan (GAG) chain - mostly sugar 95%
found in cartiladge and ecm for cushioning and lubrication
mucins
O glycosylated proteins 80% carbohydrates
create mucus
glucosamine?
amino NH2 group added to carbon 2 of glucose
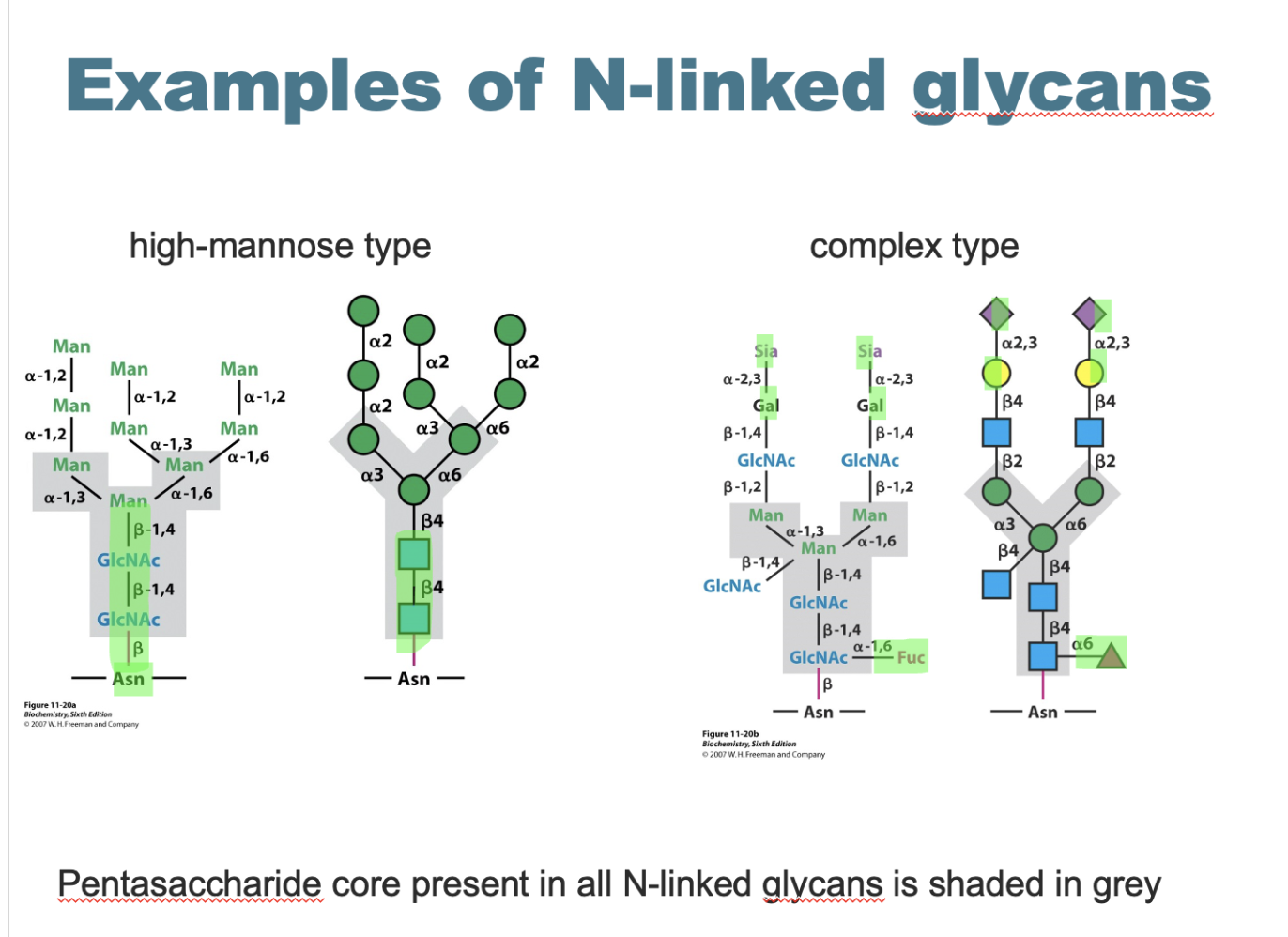
n linked glycan structure and types
main structure - 2GLcNAc and 3 mannose petasacharride core
high mannose type - many additional mannoses
complex type - extra GLcNAc, galactose, sialic acid and fucose
what are glycerophospholipids

glycerol + 2 fatty acids + phosphate + head group - amphiphatic
†he main componens of membranes
what are the functions of each glycerophospholipid

sphingomylein and cholesterol lipids function

fatty acids
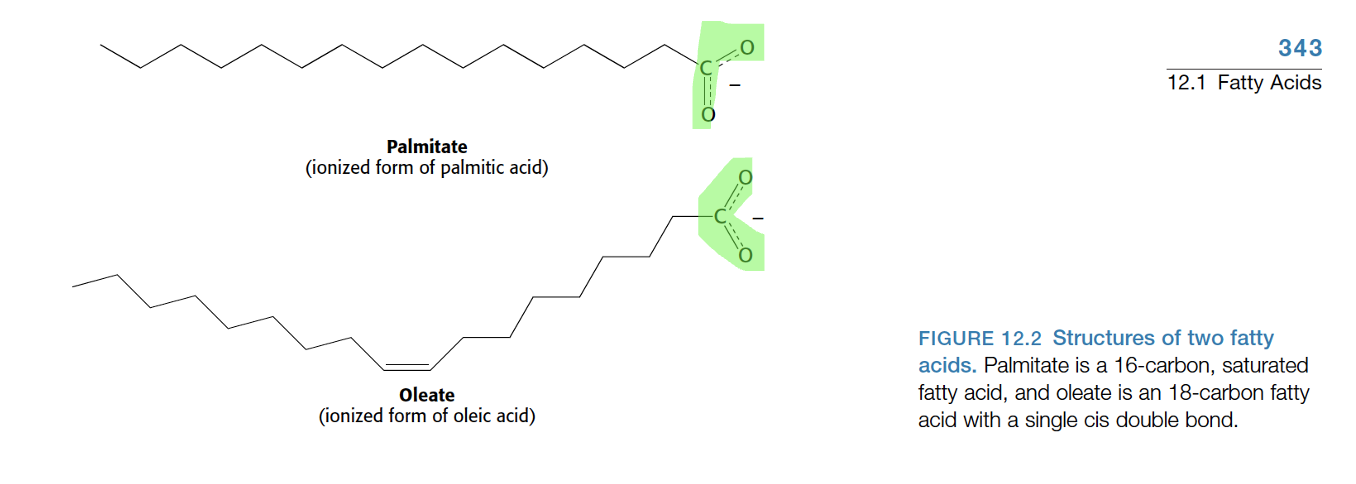
long hydrophobic hydrocarbon chain attatched to carboxiyl acid (COOH-) hydrophilic head
saturated - single bonds , tight packing and solid at room temp
unsaturated (mono,poly) - cisdouble bonds , loose packing → INCREASED MEMBRANE FLUIDITY
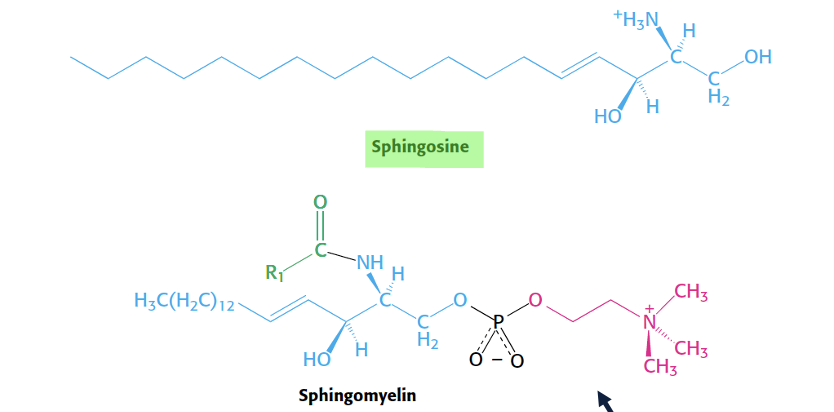
shingolipid structure
sphingosine basis - long chain amino alcohol → unsaturated hydrocarbon chain with alcohol and amino group to attavh another fatty acid
sphingosine + fatty acid = ceramide → building block for sphingolipids
sphingomylien structure and use
ceramide + phosphocoline head = sphingomylien
hydrophic tail (sphingosine + fatty acid chain at amino site)
phosphocholine polar head
major membrane lipid in mylin sheath
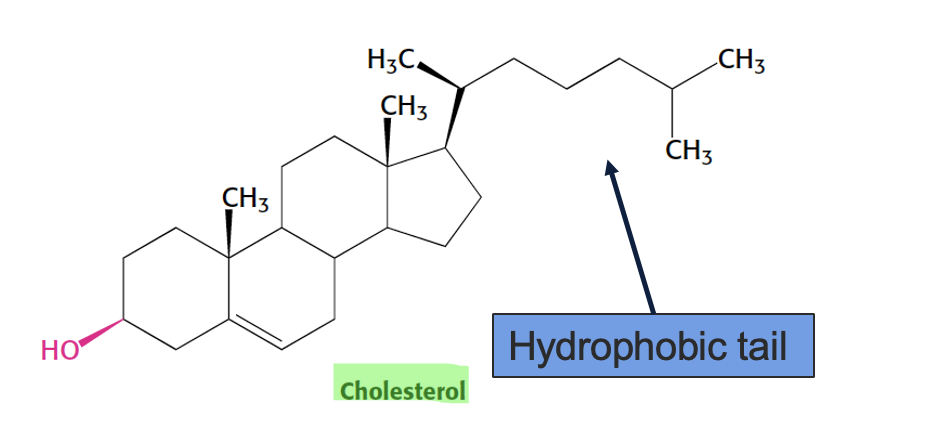
cholesterol structure and function
a sterol → 4 fused hydrocarbon rings with OH head and hydrocabon tail
amphiphatic - OH interacts with membrane surface and tail inserts into lipid bilayer
regulates membrane fluidity - at high temp → stabilizes membrane (less fluid), at low temp → prevents packing (more fluid)
bacteriorhodopsin
transmembrane protein of a-helices that span hydrophobic region of membrane
how does phosphatidylinositol (PI) signal
phosphorylated by kinase at various points
how are glycerphosphopipids synthesised, what can phosphoditate synthesis?
in ER
glycerol 3 phosphate (formed from DHAP or glycerol in liver) binds activated (saturated) fatty acid (R1-CO-CoA) at carbon 1 → lysophosphatidate
second (unsaturated) fatty acid (R₂-CO-CoA) is added to carbon 2 → phosphaditate (phosphatidic acid) PA
FATES OF PA
(DAG) a second messenger → phosphatidic acid phosphatetase (PAP) hydrolyses PA into DAG (diacylglycerol) + phosphate group (removes phosphate)
then
ON ER → triacylglycerol synthesis from DAG → add fatty acyl COA via Diacylglcerol acyl-transferase = TAG stored in fat/ liver
OR → glycerophospholipid syntheiss from PA → CTP activates PA = CDP-DAG → phosphatidylinositol/glycerol (PI/PG)
OR CTP activates head group (choline/ethanolamine) → transferring to DAG = PC/PE phosphatidylcholine/ethanolamine
Respiratory distress syndrome
lack of PC on lungs fucks surfuctant (surface tension) of fluid that keeps aveoli open aloowing gas exchange → low PC means aveoli collapse meaning fucked breathing / blue fingers
how are sphingolipids modified by sugars
ceramides → form cerebrosides by adding polar sugar head group eg glucose to UDP (uridine triphosphate)
→ gangliosides adding another sugar
Gangliosides function
-important cell surface molecules Highly prevalent in nervous tissue
tay sachs
Inherited disorder which affects motor function, then vision, fatal by 3 yr
→ unable to degrade gangliosides in lysosomes
how does ceramide form sphingosine then spingosine 1 phosphate
ceramidease adds fatty acid to ceremide amino group forming sphinogsine
sphingosine kinase adds phospahte group → spingosine 1 phosphate
cholesterol biosynthesis step 1
acetoacetyl-CoA + acetyl- CoA → formation of HMG-CoA
HMG-CoA → mevalonate catalysed by HMG-CoA reductase in RATE LIMITING STEP
reductase converts 2NADPH → 2NADP+ and release CoA in process
step 2 cholesterol biosynthesis
malanovate is phosphorylated 3 times → decarboxylated forming Isopentenyl pyrophosphate (IPP)
→ IPP condenses to form squalene C5 → C10 → C15 → C30 (IPP→GPP→FPP→squalene)
→ squalene cyclises into lanosterol which is processed to cholesterol via removal of 3 methyl groups and double bond shifting
where does cholesterol synthesis?
LIVER/intestines
4 ways HMG-CoA is contolled?
rate of mRNA synthesis → when cholesterol is low SREBP (Sterol Regulatory Element–Binding Protein) (transcription facotr) enter nucleus and increases HMG-CoA production and vice versa when cholesterol is high
rate of translation → Translation of HMG-CoA reductase mRNA is inhibited by high levels of mevalonate and dietary cholesterol.
regulation by protein degredation → High cholesterol causes the HMG-CoA to bind to Insig proteins in the ER. This marks the enzyme for ubiquitin-mediated proteasomal degradation.
regulation by phosphorylation → Phosphorylation of HMG-CoA by AMPK stops cholesterol synthesis → dephosphorylation by glucagon activates HMG-CoA
what 2 forms of cholesterol are there
free cholesterol or in an esterified form in which it is linked to long-chain fatty acids
how is cholesterol transported in the body
lipoprotein particles - hydrophobic lipid core, surrounded by polar lipids and proteins
HDL (high density lipoprotein) = ‘good’ cholesterol
LDL (low density lipoprotein) = ‘bad’ cholesterol
high LDL?
increased risk of plaque formation in vessels and atherosclerosis.
bile salts - derivitive of cholesterol
Detergents (solubilise dietary lipids)
Synthesised in the liver - glycocholate, taurocholate
Stored in the gall bladder and released into small intestine
derivitive of cholesterol - steroidsd
Progestagens - fertilisation
Glucocorticoids Mineralocorticoids
Androgens – testosterone, progesterone
Oestogens
Derivatives of cholesterol – Vitamin D
vitimin D (cholecalciferol) → additon of 2 alcohol groups makes transcription facotr - calcitriol
statins
HMG-CoA inhibitors → aim to reduce cholesterol
what does amalase, lipase, and trpsin absorb

anerobic glycolysis - how is NAD regenerated
glucose is phosphorylated twice → fructose 1,6-bisphosphate
fructose cleaved into → dihydroxyacetone phosphate and glyceraldehyde 3 phosphate
glyceraldehyeide 3 phosphate → oxidised by GAPDH where 2NAD→2NADH then phosphoryliated twice where 2ADP→ 4ATP forming 2pyruvate PER GLUCOSE
LAck of NAD stops the oxidation step so regenerates by pyruvate +NADH → Lactate and NAD via lactate dehydrogenase
sites of control in glycolysis
-Hexokinase - glucose → glucose 6 phosphate (inhibited by its high conc glucose 6-phosphate)
- Phosphofructokinase - fructose 6 → fructose 1-6 (committed step; inhibition by: high ATP, low pH, citrate;
activated by: AMP and fructose 2,6-bisphosphate)
- Pyruvate kinase PEP → pyruvate (ATP and alanine inhibit; fructose 1,6-bisphosphate activates)
phosphofructokinase 2 - regulated by?
bifunctional enzyme responsible for the synthesis and hydrolysis of
fructose 2,6-bisphosphate
has a kinase and phosphatase region - regulated by serine 460 by
protein kinase A
lactate - cori cycle
erithylocites lack mitochondria → no oxygen → pyruvate converted to lactate which muscles cant use → oxygen debt that needs cleared in liver with 6ATP (lactate→pyruvate→ glucose)
pyruvate oxidative decarboxylation
pyruvate → acetyl - COA (for tca cycle) by PDC (pyruvate dehydrogenase complex) in mitochondria
3 carbons → 2 carbons + SCoA + CO2
NAD → NADH
glycogen synthesis
IN LIVER AND MUSLCE
INITIATION - glycogenin (a glycosyl transferase) acts as primer
→ Glycogenin binds glucose from UDP-glucose to a hydroxyl group of tyrosine 194 via a-1-4 linkages
ELONGATION - glycoge synthase (GS) is phosphorylated by protein kinase A
and glycogen synthase kinase 3 (GSK3) converting from a active form to b inactive form
(however, b is still active when a high level of the allosteric activator glucose 6-phosphate is present)
→ GS add branches to an existing chain of at least four glucose residues via a-1-6 linkages
UDP gluocse formation? - whay makes this irreversible ?
glucose-1-phosphate + UTP → UDP-glucose + 2 Pi
Spontaneous hydrolysis of the ~P bond in pyrophosphate (PPi (P-P))
importanc of branching ?
increased solubility of free ends in water
increases terminal residues - the sites of action of glycogen
phosphorylase and glycogen synthase
so increases synthesis and degredation of glycogen
fatty acid synthesis
decarboxylation reaction (releases CO2) in cytoplasm via acetyl CoA using ATP and NADPH adding 2 carbons at a time
acetyl CoA moves form mitochondria to cytoplasim by converting to citrate → acetyl coA
acetyl CoA +HCO3 → malonyl-CoA via acetyl coa carboxylase (ACC) using ATP → malonyl provides 2 carbon building blocks - irreversible
Transfer to acyl carrier protein (ACP) builds chain → +2C → REDUCE via NADPH → DEHYDRATW → REDUCE VIA NADPH resulting in Palmitate (16C)
what are fatty acids sotred as
as triacylglycerides (TG).
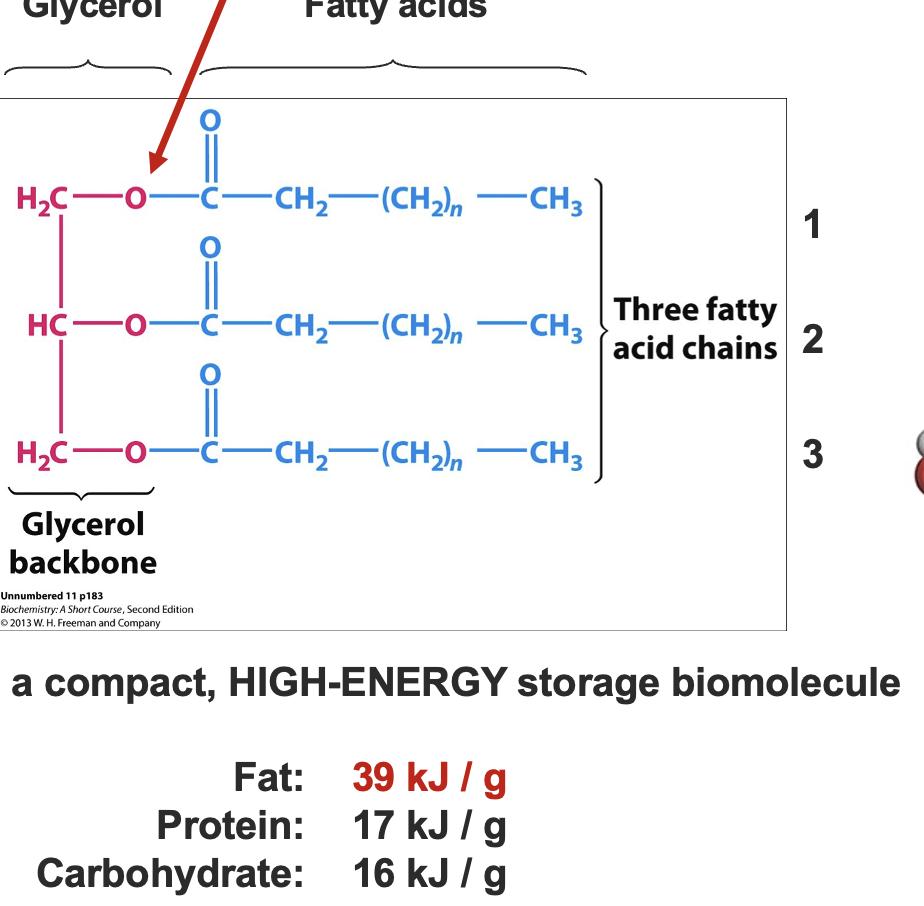
glycerol 3 phosphate + 3 fatty acids
Activation of fatty acids by CoA
Before TG synthesis, each fatty acid needs to be activated:
Thiol group of CoA → high energy thioester bond with COOH group of the fatty acid. - via Acyl CoA synthetase
Reaction driven by ATP (2 high energy bonds used).
Making TG requires high energy investment.
what vitimins need fat , what polyunsaturated fatty acids need fat
A D E K
linoleic acid [C18:2] and linolenic acid [C18:3].
why do lipids (fatty acids ) need lypoproteins to be transported
they are hydrophobic and insoluble in aqueous
environments.
fatty acid oxidation (b-oxidation)
activation - thiol of coenzyme A high energy thioester bond
with the carboxylic acid group of the fatty acid. - via Acyl CoA synthetase using ATP and hydrolysing pyrophosphatetransport - Carnitine is an acyl-carrier that transports fatty acids into mitochondria
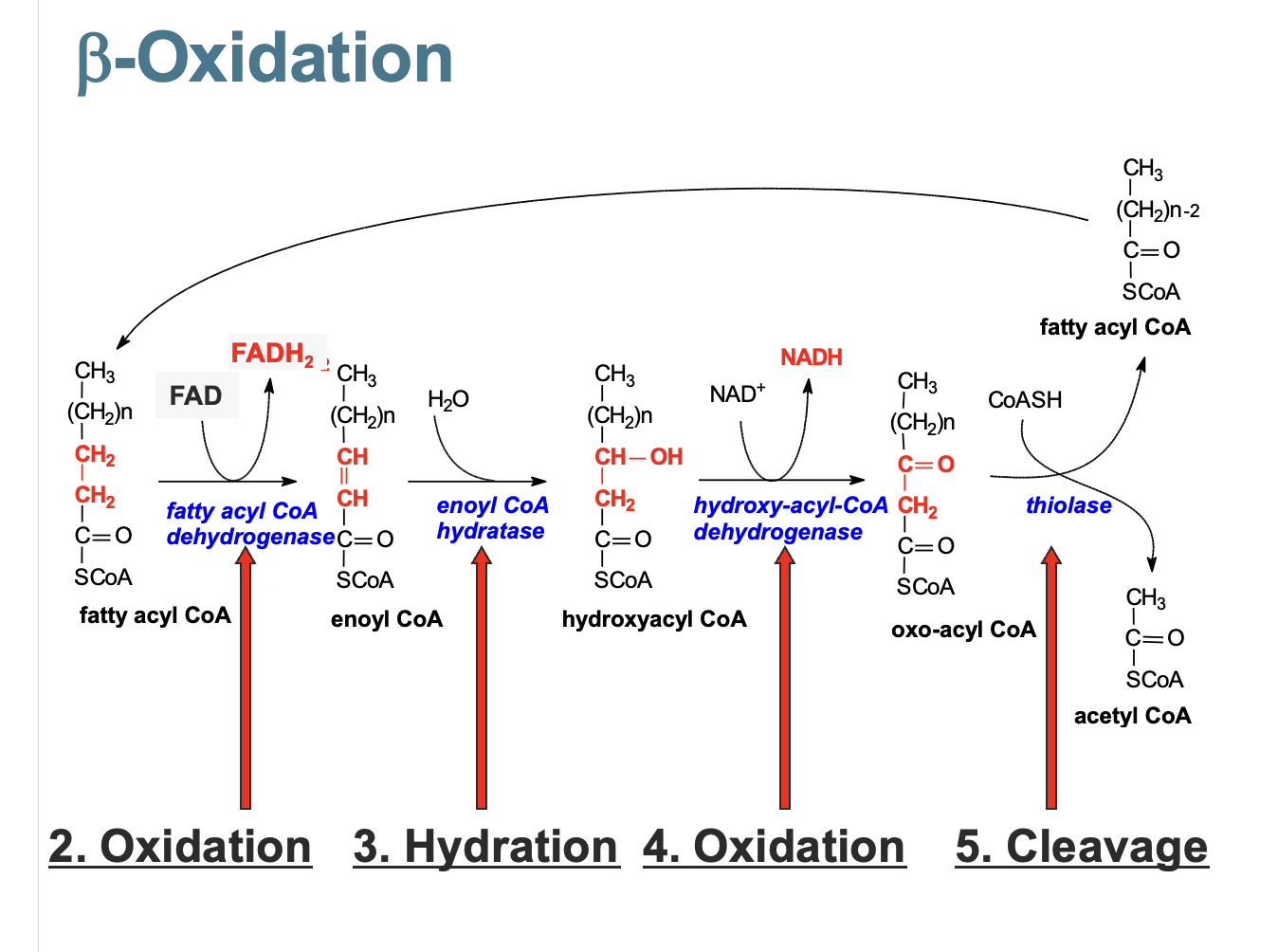
double bond created
water added to double bond
B oh → ketone group
ketone group attacked by coA splitting into fatty acyl coA and acetyl CoA
degredation of unsaturated fatty acids
double bond breaking requires - cis-D3-Enoyl CoA isomerase PLUS
2,4-Dienoyl CoA reductase
odd chain fatty acids
propionyl CoA → succinyl CoA via bicarbonate
gluconeogenesis - making glucose
Conversion of pyruvate into glucose (mainly in liver).
Major noncarbohydrate precursors are:
lactate, propionate, amino acids, glycerol
Where do they come from ?
- lactate, rate of glycolysis exceeds the rate of
oxidative metabolism
- amino acids, breakdown of proteins
- propionate and glycerol, hydrolysis of triacylglycerols

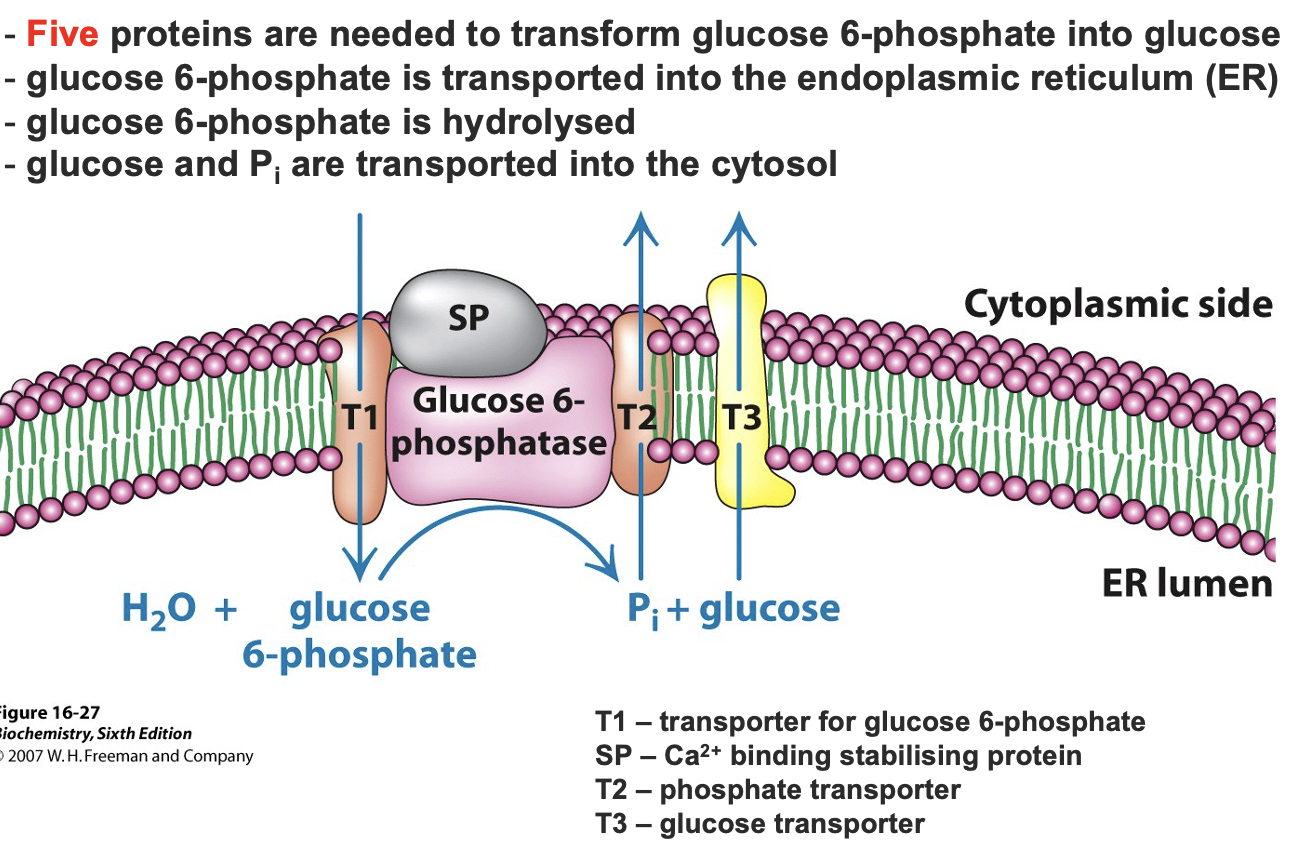
glucose 6 phosphateinto glucose
glucose 6 phate → gluose via Glucose-6-phosphatase removing the phosphate group in liver and kidneys in the ER
fructose 1-6 biphosphate
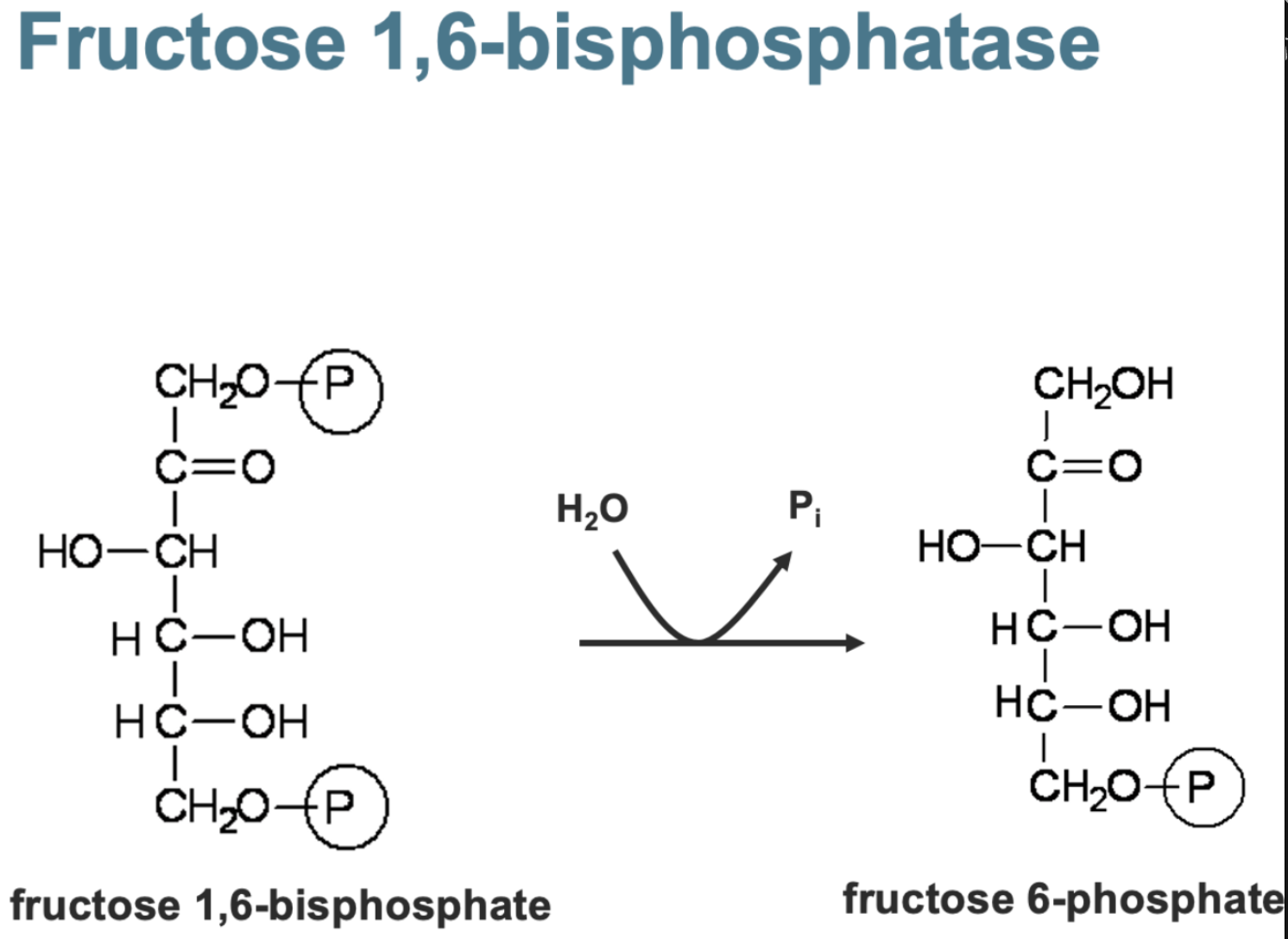
fructose 1-6 phosphate — tetramer with mg /zn /mn as cofactors
AMP - low energy signal turns off converison
Fructose 2-6 phosphate - strong inhibitor activating glycolysis instead
citrate - activator
pyruvate converison
mitochondira 1. Pyruvate carboxylase activated by acetyl coA converts pyruvate → oxaloacetate (OAA) using ATP (adds CO2)
cytosol 2. Phosphoenolpyruvate Carboxykinase (PEPCK) - converts OOA → PEP + CO2 by adding phosphate using GTP
OAA in mitochondria can be reduce to malate or transaminated to aspartate to move into cytosol
transamination
transfer of amino group of amino acid to an a-ketoacid
Decarboxylations often drive reactions
that are otherwise highly endothermic !!!