Cell Bio Review
1/73
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
74 Terms
A glucose molecule is to starch as
a nucleotide is to a nucleic acid
a shortage of phosphorus in the soil would make it especially difficult for a plant to manufacture
DNA
Lipids differ from other macromolecules in that they
are not true polymers
unsaturated fats
have double bonds in their fatty acid chains
which of the following is an organic molecule
a. o2
b. h20
c. hcl
d. ch4
CH4
monomers are attached together to create polymers when a hydroxyl group and a hydrogen atom are ___ in a _____
added; dehydration synthesis
which of these is not a lipid?
a. steroid
b. fat
c. polysaccharide
d. wax
polysaccharide
animals store their excess carbohydrates in the form of
glycogen
nearly all _____ are __________
enzymes; proteins
the difference between one amino acid and another is found in the
R group
which of the following is not a type of RNA?
a. nRNA
b. mRNA
c. rRNA
d. tRNA
nRNA
each amino acid in a polypeptide is specified by
a codon
which of the following statements is correct about the genetic code?
a. each codon encodes an amino acid
b. each amino acid is encoded by only one codon
c. a codon consists of three nucleotides
d. a codon and its complementary anticodon have the same sequences
C.
codons code for trinucleotides, amino acids are not encoded by codons, codon and anti are different
the process of obtaining a copy of the information in a gene as a strong of messenger RNA is called
transcription
the site where RNA polymerase attaches to the DNA molecules to start the formation of an RNA molecules is called
promoter
the process of taking the information on a strand of messenger RNA and building an amino acid chain, which will become all or part of a protein molecule, is called
translation
if an mRNA codon reads UAC, its complementary anticodon will be
ATG
Which of the following accurately describes gene expression in prokaryotic cells?
a. all genes are on all the time in all cells, making the needed amino acid sequences
b. some genes are always off unless a promoter turns them on
c. some genes are always on unless a promoter turns them off
d. some genes remain off as long as a repressor is bound
D
which of the following statements is correct about eukaryotic gene expression?
a. mRNAs must have introns spliced out
b. mRNAs contain the transcript of only one gene
c. enhancers act from a distance.
d. some genes remain off as long as a repressor is bound
C
which of the following is not a mechanism of controlling gene expression in eukaryotic cells
a. blocking translation with small interfering RNA
b. activating an enhancer
c. translating a gene as it is being transcribed
d. alternative splicing of the primary RNA transcript
B
dehydration synthesis
molecules form by removing water
hydrolysis
molecules bread down by adding water
primary structure
sequence of its amino acids
secondary structure
formed by hydrogen bonds between nonadjacent carboxyl and amino groups
tertiary structure
results from interactions between R-groups
quaternary structure
results from hydrogen and ionic bonds between separate tertiary structures
X ray crystallography
solid crystals of purified protein are placed in an X-ray beam, and the pattern of deflected X rays is used to predict the positions of the atoms within the protein crystal
five different nitrogenous bases
adenin, guanine, cytosine, thymine (DNA), uracil (RNA)
difference between DNA and RNA structure
OH or H in sugar base
U or T
single strange or double strand
difference between DNA and RNA function
RNA transfers the genetic information from DNA to the protein-making machinery
DNA stores the genetic information
A and T can align to form _____
two hydrogen bond
G and C can align to form ______
three hydrogen bonds
carbohydrate
has a C,H,O usually 2:1 (Hydrogen:Oxygen)
lipids
not water soluble, made of fatty acid chains that store energy
fat → triglyceride
saturated fats
each carbon in the hydrocarbon chain is bound to two hydrogen atoms
tightly packed, solid at room temp
unsaturated fatty acids
at least one carbon in the hydrocarbon chain is bound to just one hydrogen
loosely packed, liquid at room temp]
central dogma
replication → DNA → transcription → RNA → translation → protein
DNA replication
process by which DNA makes a copy of itself during cell division
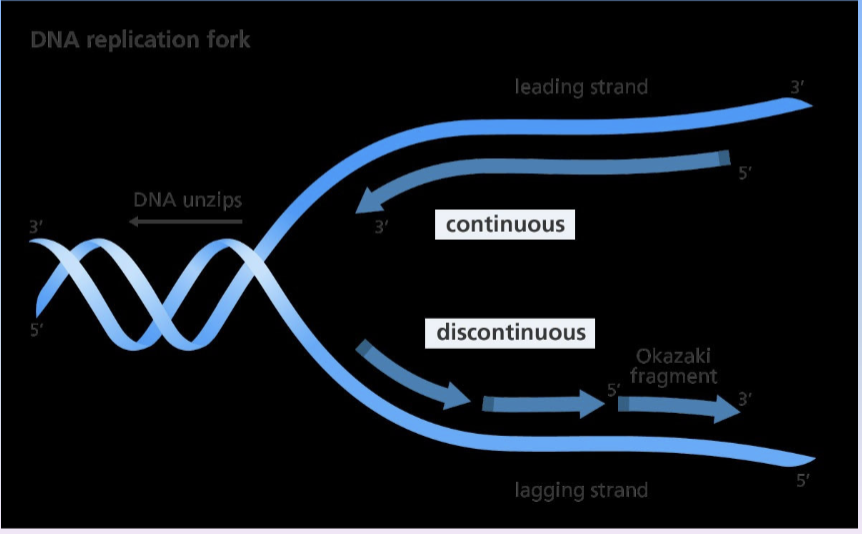
replication fork formation
primer binding
elongation
termination
replication fork formation
process of DNA replication where it is unzipped into two single strands via DNA helices, an enzyme that disrupts the hydrogen bonding between base pairs
only processes in the 5’ to 3’ direction
5’ has a phosphate group attached and 3’ has a hydroxyl group attached
but, it is bidirectional so 3’ to 5’ is the leading strand and 5’ to 3’ is the lagging strand.
primer binding
in DNA replication, a short piece of RNA that will bind to the 3’ end of the strand (the starting point for replication)
generated by the enzyme DNA primase
elongation
process during DNA replication where DNA polymerases will create the new strand
DNA pol III is the main replication enzyme while DNA polymerases I, II, IV, V, are responsible for error checking and repair (prokaryotic cells)
In eukaryotic cells, there are DNA pol alpha, delta, epsilon
replication proceeds in the 5’ to 3’ direction on the leading strand, the newly formed strand is continuous.
lagging strand will bing with the primers, DNA poly adds Okazaki fragments, leading to a discontinuous replication because of disjointment
termination (DNA rep)
continuous and discontinuous strand have been formed, an exonuclease removes all RNA primers and replaced with the bases, another exonuclease proofreads
DNA ligase joins Okazaki fragments together forming a single unified strand.
ends of each strand has telomeres, repeated sequences of DNA that act as protective caps
telomerase, an enzyme, will catalyze the synthesis of telomere sequences at the end of the DNA
topoisomerase or DNA gyrase
unwinds and rewinds DNA strand to prevent the DNA from becoming tangled or supercoiled
point mutations
a single base substitution
deletion
a small DNA segment is lost
insertion
a segment of DNA is added
frame-shift mutation
modification of the reading frame after a deletion or insertion, resulting in all codons downstream being different
gene expession
the process where gene info is used to make a function gene product that will make protein as the end product
gene expression in prokaryotes
a gene is translated as it is transcibed
no nucleus, so newly made mRNA is directly accessible to ribosomes
no introns, no splicing of mRNA
gene expression in eukaryotes
a nuclear membrane separates the process of transcription and translation
mRNA has to move to the cytoplasm after splicing
steps:
RNA pol transcribes RNA from DNA
Introns are excised, exons are spliced together to from mRNA
mRNA out of nucleus to cytoplasm where ribosomal subunits bind to it
tRNA molecules attach to amino acids and go to ribosome
tRNA + amino acids → A site
peptide bonds form in P site and tRNAs exit at the E site
grows until polypeptide is complete
transcription
the process of creating a complementary RNA copy of a sequence of DNA
steps:
initiation: RNA pol binds to promoter of a gene and begins to unwind
elongation: RNA pol reads 3’ to 5’ and adds complementary ribonucleotides
termination: RNA pol hits stop signal or falls off
translation
mRNA from transcription is decoded by the ribosome to make an amino acid chain or polypeptide that will be a proteinc
degeneracy
one amino acid has multiple codons
each codon specified the amino acid to placed at the corresponding position along a polypeptide
mRNA
carries info on AA sequences of proteins from DNA to ribosomes
tRNA
serves as a translator molecule in protein synthesis; mRNA codons → amino acids
rRNA
catalytic roles and structural roles in ribosome
primary transcript
precursor to mRNA, rRNA, tRNA before being processed, can catalyze its own splicing
snRNA (small nuclear)
structural catalytic role in spliceosomes, protein and RNA that splice pre-mRNA
ribsome
A,P,E sites important for gene expression, two subunits
steps in translation
mRNA leaves nucleus and goes to ribsomes
mRNA brings small subunit
tRNA bring amino acid to ribosome and anticodon tRNA binds to codon mRNA
amino acids bind and form polypeptide
free tRNA is released
other tRNAs bring amino acids to the ribosome
cl
class 2 GFP
GFP with a phenolate anion in the chromophore (EX: EGFP [Enhanced Green Fluorescent Protein])
The most common mutation to cause ionization of the phenol of the chromophore is when Ser65 is replaced by Thr (mutation S65T)
These mutations are set to improve the folding efficiency, not to increase the brightness (even though this could be an indirect effect).
class 3 GFP
GFP with a neutral phenol in the chromophore
When T203I largely suppresses the 475 nm peak, there is a singular peak at 399 nm, meaning that the chromophore will be neutral in almost all the ground-state molecules.
which is beneficial because you can have a more specific signal as well as reduced overlap to clearly see which light is emitted/absorbed.
class 4 GFP
GFP’s chromophore’s phenolate ion has an aromatic ring stacked next to it (T203F).
YFP
class 5 GFP
GFP protein where Tyr66 is replaced with Trp (Y66W), making the chromophore have an indole instead of a phenol.
CFP (cyan)
One characteristic of these (unexplained) is that there have double-humped excitation and emission peaks
class 6 GFP
GFP protein having a His instead of a Tyr6 (Y66H), making the chromophore have an imidazole instead of a phenol
This class has a low fluorescence quantum yield and is easily susceptible to photobleaching. There is also a EBFP (Enhanced BFP) that has emission and excitation peaks close to the regular BFP, but slightly lower. Enhanced BFPs will be more photostable and have a brighter intensity.
class 7 GFP
GFP protein having Phe at 66 instead of Tyr (Tyr66 for the wild type), the mutation being Y66F
shortest wavelengths obtained for a GFP.
expression
If there are more gene copies and more promoters, there will be more protein present in the cell. It was studied that mammalian codons present will not hurt the expression levels present in other organisms, as seen with the additional valine or alanine in from of the initial methionine. In turn, GFP can be expressed freely, under the right conditions (highly specific), the protein folding autonomously. For example, after denaturing the protein, it could refold on its own into a cylindrical structure (chromophore must be in the middle, surrounded by the shell to properly function as a GFP).
formation
Folding is a process that takes place under the right conditions, and the general mechanism is that GFP folds, following by the imidazolinone forming via nucleophilic attack on the Gly67 on the residue 65, followed by dehydration, which requires oxygen. Fluorescence cannot occur without atmospheric oxygen and by dehydrating the a-b bond in residue 66, the imidazolinone is in conjunction with the aromatic group. The presence of chaperones can help the protein fold, as well as mutations present in the GFP (especially Enhanced GFPS). The more efficient the fold, the faster and more reliable the reaction will be.
maturation
Once the protein is mature, oxygen is no longer required. The oxidation appears to be the rate-limiting step in the formation of this protein. However, it has not been studied on how to utilize the oxidative properties in the GFP, but it can be hypothesized that with more oxygen added, there will not be a faster reaction (with more oxygen present, we don’t breathe faster, so why would the GFP be any different? Not sure how right this is, just taking a guess).
renaturation
Since the conditions to make GFP are super specific, the conditions to keep it active and functioning are similar. Under environmental duress, the protein may denature and unfold. Under the right conditions, if they return, the protein can refold, but must be rehydrated in order to be fully functioning.
passive GFP applications
tags and indicators, used as a tracker for what you are looking for
ex: C. elegans expression pattern for mechanosensory neurons
active GFP applications
FRET, calcium sensitivity, dimerization
FRET
Fluorescence Resonance Energy Transfer (FRET) is a technique that takes two fluorophores with an overlapping emission (donor) and excitation (acceptor) spectrum. Highly dependent on distance, need to be close distance (<100 A).
o One method of FRET is protease action where a BFP (class 6) is fused to GFP (class 2). The donor BFP has an emission spectrum peaking at 447 nm and it overlaps with the excitation spectrum of the GFP, peaking at 489 nm.
o Disadvantage: high background signal that struggles to detect trace interactions in early forming cellular components.