Genetics Final Exam
1/85
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
86 Terms
Most Common Cancers
US: Skin Cancer
World: Lung Cancer
G1/S Checkpoint
Cell monitors size and DNA integrity
Loss: gene amplification, chromosomal rearrangements, lack of apoptosis
G2/M Checkpoint
Cell monitors DNA synthesis and damage
Loss: translocations
M Checkpoint
Cell monitors spindle formation and attachment to kinetochores
Loss: polyploidy and aneuploidy
Cyclins and Kinases
Control passage through cell cycle
Cancer Driver Genes
Tumor Suppressor Genes, Oncogenes, DNA Damage Response (DDR) genes
Tumor Suppressor Genes
E.g. BRCA1 and BRCA2
LOF, recessive
Oncogenes
Ras (HRAS, KRAS, NRAS)
GOF, dominant
DNA Damage Response (DDR) genes
E.g. ARID1A
Usually recessive
Sustaining proliferative signaling
Cancer cell stimulate own growth without external signals
Evading growth suppressors
Cancer cells ignore tumor suppressor genes
Resisting cell death
Avoid apoptosis
Enabling replicative immortality
Cancer cells activate telomerase and become undifferentiated
Genome Instability and Mutation
Increased mutation rates
Non-mutational Epigenetic Reprogramming
Epigenetics change
PARP1 in BRCA1/2 Cancer
PARP1 is DDR protein, and PARPi causes synthetic lethality by stopping PARP1 from repairing small damages
Synthetic Lethality
One copy of an allele is viable but two copies leads to cell death
Organismal Hallmarks of Aging
Loss of skin elasticity, Hair loss/greying, Menopause, Cognitive impairment, Muscle strength,
Age-related diseases: lung disease, stroke, cardiovascular disease, diabetes, kidney failure, osteoarthritis, cancer, dementia & Alzheimer’s
Cellular/Molecular Hallmarks of Aging
Altered intercellular communication
Stem cell exhaustion
Cellular senescence
Mitochondrial dysfunction
Deregulated nutrient-sensing
Loss of proteostasis
Epigenetic alterations
Telomere attrition
Genomic instability
Mitochondrial Dysfunction (with Aging)
Susceptible to mutations which affects efficiency of oxidative phosphorylation (making ATP) and increasings ROS (reactive oxygen species)
DNA Damage (with Aging)
Genomic instability, Stem cell exhaustion, Cellular senescence, Altered intercellular communication
Unrepaired DNA damage causes cells to sensece or undergo apoptosis, reduced stem cell regeneration, and disease
Telomere attrition (with Aging)
When telomeres get too short, cells undergo growth arrest or apoptosis—non-immortal cells
Generally longer=healthier, but mice and some human cancers are the exception
Associated with microplastics
Loss of protein homeostasis (with Aging)
Free radicals react with some amino acids (histidine, arginine, lysine, proline and methionine) to impact protein function. Proteins can also react with glucose, impacting the protein’s structure and function.
Epigenetic alterations (with Aging)
DNA methylation decreases with age except for CpG sites. Under-methylating leads to expression of repeat DNA.
Deregulated nutrient-sensing (with Aging)
Reducing caloric intake prevents diseases related to impaired metabolism.
Measuring Biological Age
Telomere length, epigenetic clocks, blood biomarkers, physiological tests (muscle strength, memory decline, etc)
Horvath’s Epigenetic Clock
Gathered methylation data from 7k people and found 353 age-related CpG sites to estimate biological age and chronological age. Sperm had very low biological age and cancer cells were sporadic
DNAm PhenoAge
Compared DNA 513 methylation sites with clinical biomarkers. Better than Horvath’s for disease/mortality prediction
Factors affecting biological aging
Physical Activity, Diet, Socioeconomic status, Education level, Smoking/Drinking, Metabolism (BMI, Diabetes), Menopause, Infections (HIV), Heart/Lung Function, Cancer, Male, Neuropsychiatric (PTSD, Depression, Schizophrenia, Alzheimer’s Parkinson’s)
APOE
Fat-binding protein that mediates cholesterol metabolism—associated with extreme longevity alongside lncRNA
CIRBP
Protein expressed in bowhead whales that increases NHEJ w/ more accuracy—extends life
Evolutionary Theories of Aging
Mutation accumulation theory, Antagonistic pleiotropy theory, Disposable soma theory
Mutation accumulation theory
Natural selection is blind to variants that impact organism AFTER reproduction
Antagonistic pleiotropy theory
Natural selection choose traits are beneficial when young and harmful when old (cell senescence: when young helps in development, wound healing, cancer prevention)
Disposable soma theory
Natural selection has limited energy budget between reproduction, growth, and aging
Cellular Potency
Ability for cells to become lots of specialized cells
Determined by DNA methylation, PcG and TrxG (group proteins that control epigenetics)
Totipotent
Cells become ANY cell type (Zygote starts are this)
Open genome: global DNA demethylation
Pluripotent
Cells become MANY cell types
No X-inactivation, genes for all differentiation off (by PcG), Promoter hypomethylated
Multipotent
Cells become MORE THAN ONE cell type
X-inactivation, lineage-specific genes for differentiation off (by PcG), Promoter hypermethylated
Unipotent
Cells are STUCK as one cell type
X-inactivation, PcG silences all differentiation, TrxG turns on lineage-specific genes, Promoter hypermethylated
PcG/TrX
PcG:PRC2 writes H3K27me3 in pluripotent cells, PRC1 causes chromatin to condense in multi/unipotent cells
TrX: Unipotent cells have genes turned on by Trx writing H3K4me3
Hox Loci
Regulation body development, directed by morphogens
Control Development
TFs, Pioneer TFs, and RNA (non mRNA)
Phase 0
10 to 15 patients
Study pharmacodynamics and pharmacokinetics
Discovery Science
Studies in lab, including animal studies
Phase I
20 to 100 patients
Study in small amount of patients to determine maximum tolerable dose
single-ascending: different patients get higher amounts
multiple-ascending: each patients get more small doses
Phase II
50 to 100 patients
Study in more patients for drug effecacy (and more safety!)
Phase III
300 to 3000+ patients
Confirms safety/efficacy, but also for side effects and to compare to current treatments
Phase IV
In the Market
FDA-approved drugs are surveilled for long-term and rare effects, especially with new data groups
Pharmacodynamics
Study of the biochemical and physiological effects of drugs and their mechanisms of action (drug’s impact on the body)
Pharmacokinetics
Study of the absorption, distribution, metabolism, and excretion of drugs (often referred to as ADME) (body’s impact on a drug’s activity)
Pharmacogenomics
Study of how patients’ genomes impact their response to drugs
Personalized medicine
Tailoring patient care based on genomic sequence, environmental exposures/lifestyle, precise disease classification, personal/family health histories
Pharmacology
Interaction of chemicals with biological systems to yield therapeutic or other beneficial effects
Toxicology
Interaction of chemicals with biological systems to yield adverse effects
Where Drugs are metabolized (altered)
The liver (bioactivates and excretes)
CYP enzymes
Oxidize hydrophobic drug centers to be more hydrophilic to leave body, sometimes makes drugs more bioactive instead of eliminating
We have 57 and variants affect drug metabolism
ED50
Dose where 50% of patients benefit
TD50
Dose where 50% of patients experience toxic side effects
Therapeutic range
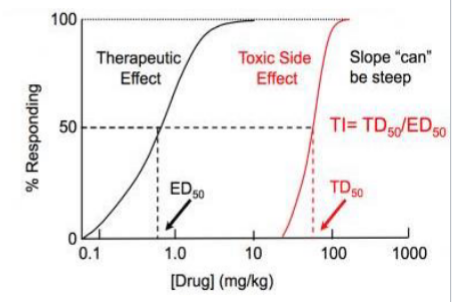
Therapeutic Index (TI)
TD50/ED50, want a large TI
Personalized Medicine Examples
CarT Therapy (cells recognize proteins only in cancer cells), Chemotherapy (choosing drug based on type of breast cancer ER/PR/HER2), Antibiotics (choosing drug bacteria isn’t resistant to), Diet (choosing food based on glycemic index)
Transposable Elements (TEs)
segments of DNA that can move to different locations
Most inactive (less than 0.05%)
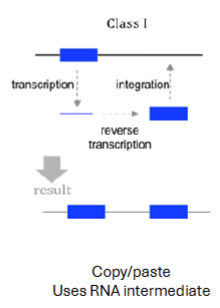
Class I/Retrotransposons
Copy themselves into RNA then to DNA and then paste into a new location
~40-50% of human genome
LINE —> mRNA —> some protein (transposase and integrase) some mRNA —> transposase protein turns mRNA to cDNA and integrase protein inserts into genome
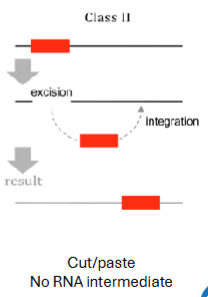
Class II/DNA transposons
Can just cut and paste themselves
~3% of human genome
Transposase binds to transposon, makes loop that cuts it out, inserts loop elsewhere, loop flattens out (gene for transposase on transposon), replicates by jumping during replication
Can’t jump anymore
Transposon Hypothesis
Theory that introns evolved from transposon jumping into coding sequences
Retrovirus
Transcribed into cDNA that’s inserted into chromosomes, e.g. syncytin proteins that made placenta for mother/child nutrient transfer and supressyn which binds to receptors to block invaders—common in placenta to protect germline
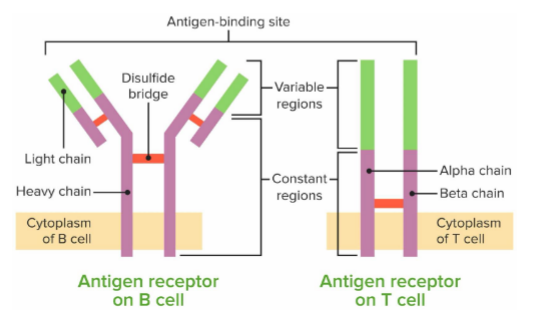
B and T cell receptors
do NOT have the same DNA as rest of cells
bind to antigens with high specificity
B+T cells replicate really quickly when infection happens to launch attach, and body remembers the activated cells for future infections
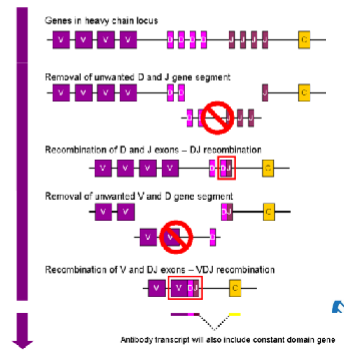
VDJ gene regions
Somatic recombination
D and J joins (everything between deleted)
V and D join (everything between deleted)
extra C and V are spliced out
Recombination Signal Sequences (RSS)
Flank individual V, D, and J regions
Allows Rag1 and Rag2 proteins to bind for cleavage
NHEJ mechanism is used to ligate ends together, slight variability each time
Affinity Maturation
Rapid replication with high mutation rate, ensure different levels of affinity to invader, highest affinity binds are replicated again
Somatic Hypermutation
Mechanism of affinity maturation
When BCR binds to antigen, AID deaminates C (turns to U) which causes mutations that affects affinity
Rag transposon Hypothesis
WDJ recombination originated from transposons jumping into receptor gene
Bacteriophage lysogenic cycle
Phage DNA incorporated into host’s genome
Bacteriophage lytic cycle
Phage DNA circular and separate from host DNA
Cas9
Type II single-effector nuclease
Knockout
Permanently removes or inactivates a gene
Knockdown
Temporarily reduces gene expression
Knock-in
Inserts new genetic material
Closest living relatives
Chimpanzees
Human Lost Genes
~80, smell, hair. smaller chewing muscles (myosin MYH16)
hCONDEL
human conserved deletions that are present in other species except humans
HAR
human accelerated regions that show more differences than expected, explained by genetic grift
Closest Non-Living Relative
Denisovans and Neanderthals
Genetic Distances
Other humans: ~0.6%
Chimps: ~2%
Most human variation is between individuals, not populations
STR
Short tandem repeats used for DNA profiling (identifying individuals)