eukaryotic transcription 3
1/34
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
35 Terms
Why does post-transcriptional regulation exist in eukaryotes?
Because transcription occurs in the nucleus and translation in the cytoplasm, allowing extra regulation and quality control 🧪➡️🏭.
What must happen to eukaryotic RNA before it is exported?
It must be modified, processed, and quality-checked ✔️.
What is the C-terminal domain (CTD) of RNA polymerase II?
A tail region made of 52 repeats of a 7-amino-acid sequence that coordinates RNA processing 🧵.
Which amino acids in the CTD are phosphorylated?
Serine 2 and Serine 5 (Ser2 & Ser5) 🔋.
Why is CTD phosphorylation important?
It recruits capping enzymes, splicing factors, and 3′ processing machinery during elongation 🧲.
What are the five major post-transcriptional modifications of eukaryotic mRNA?
5′ capping 🎩, splicing ✂️, 3′ processing & polyadenylation 🧬, RNA editing ✏️ (rare), and export 🚪.
What is the structure of the 5′ cap?
A 7-methylguanosine (m⁷G) linked by a unique 5′–5′ triphosphate bond 🔗.
When is the 5′ cap added?
When the RNA is ~25–30 nucleotides long ⏱️.
Why is the 5′ cap essential?
It protects mRNA from degradation, enables nuclear export, allows translation, and marks RNA as mRNA 🛡️.
What enzymes are involved in 5′ capping?
RNA triphosphatase, guanylyltransferase, and methyltransferase 🧪.
Describe the mechanism of 5′ capping.
Step 1: Triphosphatase removes γ-phosphate ➖;
Picture:
PP——RNA
Step 2: Guanylyltransferase adds GMP (guanine) via 5′–5′ bond 🔗
G–PP——RNA
Step 3: Methyltransferase methylates guanine → m⁷G (7-methylguanosine)🎩.
m⁷G–PP——RNA 🎩
How is the 5′ capping process coordinated?
Capping enzymes bind the phosphorylated RNA Pol II CTD during transcription 🧲.
What binds the 5′ cap after it is added?
Cap-binding complexes (CBCs) 🧢.
What are the functions of cap-binding complexes?
Protect mRNA, promote export, enable pioneer translation, and allow nonsense-mediated decay 🔍.
What happens to improperly capped RNA?
It is recognised and destroyed as part of quality control 🚮.
How is faulty capping handled in yeast?
The Rai1–Rat1 complex removes faulty caps and degrades RNA 🧬.
How is faulty capping handled in mammals?
DXO removes defective caps and degrades the RNA 🧪.
Why can influenza virus not cap its own RNA?
It is a negative-sense ssRNA virus lacking capping enzymes ❌.
What is cap snatching?
Influenza steals capped RNA fragments from host mRNA to prime viral transcription 🦠.
What happens to the host mRNA during cap snatching?
It is decapped and degraded 📉.
Which drug inhibits influenza cap snatching?
Baloxavir marboxil 💊.
What type of genes do eukaryotes have?
Monocistronic genes (one gene → one mRNA) 🧬.
Why do eukaryotic genes contain introns?
To increase regulatory potential and allow multiple protein products 🔀.
On average, how many introns does a human gene contain?
About 8–9 introns 📊.
What are the three types of RNA splicing?
Nuclear pre-mRNA splicing ✂️, Group II self-splicing 🔁, Group I self-splicing 🔂.
Which type of splicing is most common?
Nuclear pre-mRNA splicing using the spliceosome 🧠.
What is the branch point nucleotide in nuclear splicing?
Adenine (A) 🔺.
What are Group II introns?
Rare self-splicing introns in organelles where RNA acts as a ribozyme 🧬.
What are Group I introns?
Very rare introns in some viruses with guanine (G) as branch point 🧪.
What are the conserved splice site sequences?
5′ splice site = GU, 3′ splice site = AG, branch point = A 📍.
What is the polypyrimidine tract?
A pyrimidine-rich sequence just upstream of the 3′ splice site 🧵.
Describe the mechanism of splicing.
Step 1: 2′-OH of branch point A attacks 5′ splice site → lariat 🔁;
Step 2: 3′-OH of exon 1 attacks 3′ splice site → exons ligated ✂️.
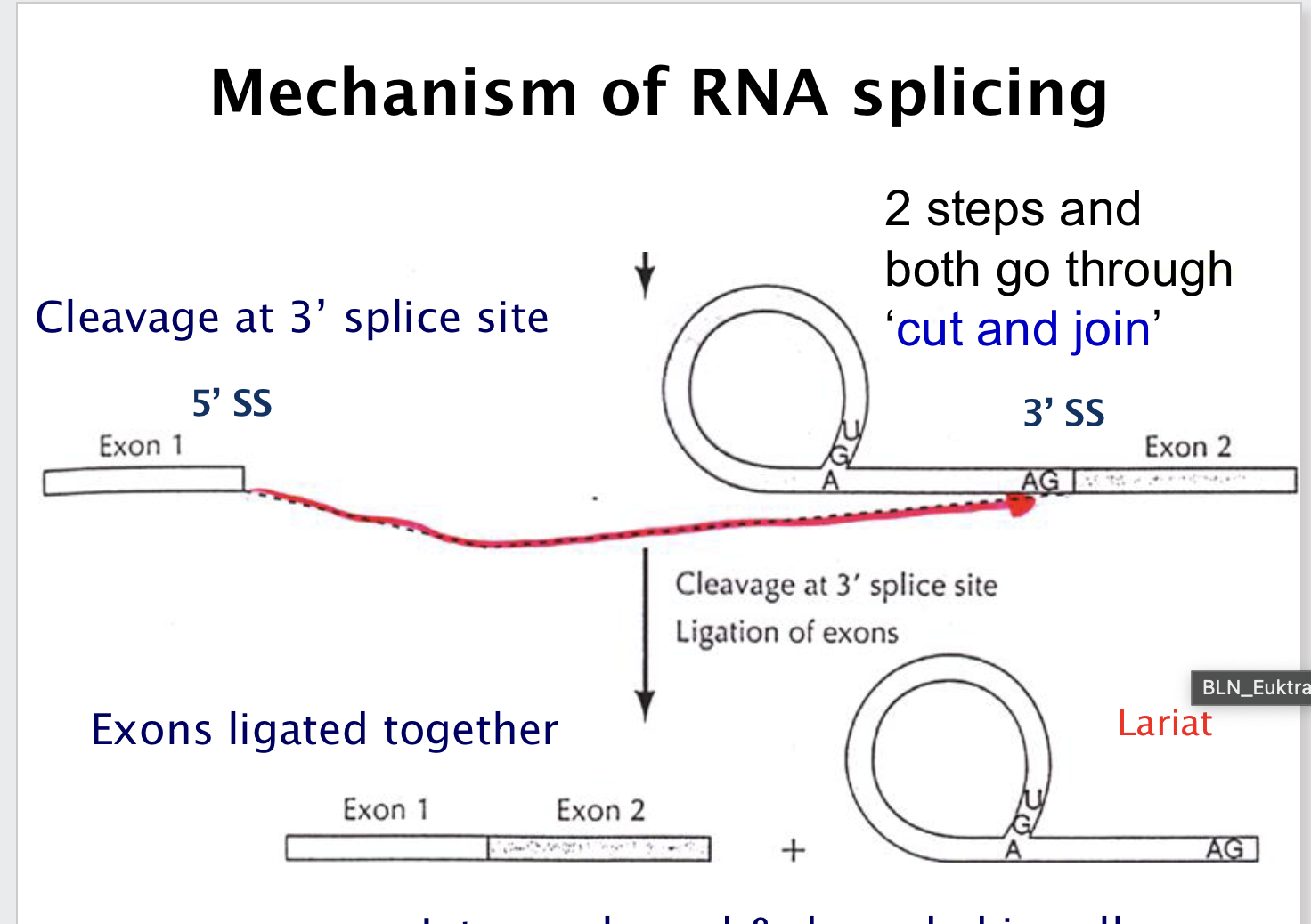
What happens to the intron after splicing?
It is released as a lariat and degraded 🚮.
What is the key takeaway about post-transcriptional regulation?
RNA processing is tightly coupled to transcription and ensures stability, accuracy, and correct gene expression 🧠.