Unit 2 Protein Function Cell Bio
1/73
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
74 Terms
reaction equilibrium
Keq= Kforward/Kreverse = [products]/[reactants] (constant)
![<p>K<sub>eq</sub>= K<sub>forward</sub>/K<sub>reverse</sub> = [products]/[reactants] (constant)</p>](https://knowt-user-attachments.s3.amazonaws.com/e0c9daba-2ee1-4d2e-b6e4-ac77790b245c.png)
binding reactions
formation of intermolecular complexes is an equilibrium reactions; molecules bounce into each other randomly; noncovalent bonds stabilize the interaction
association and dissociation rates
each binding reaction has a unique…
never zero
the dissociation rate is ….
Kon and Koff
non-covalent bonds determine these
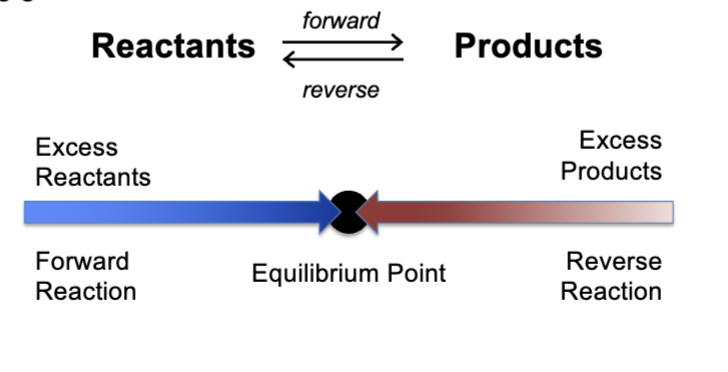
forward or reverse
every reaction can occur in the ______ or ______ directions
converge
reaction systems will _____ on their equilibrium
energy
moving reactions away from equilibrium requires ______; the further reactions are away from equilibrium, the more ______ is required or released
Gibbs Free Energy
change in this measures how much energy would be released by a reaction to occur given reagent concentrations
makes no statement about reaction rate
ΔG = 0
the reaction is at equilibrium
negative ΔG
reaction is exergonic, releases energy, and can proceed spontaneously
positive ΔG
reaction is endergonic, requires energy input, and cannot proceed spontaneously
move a system away
ΔGforward=-ΔGreverse
accounts for energy needed to ____________ from the equilibrium
absolute energy state
cannot calculate this before or after a reaction, only the change in free energy (ΔG)
how to calculate Gibb’s free energy
ΔGo’ measured under defined “standard conditions”: all products and reactants are 1 M, pH=7
ΔGo’ is specific to each reaction
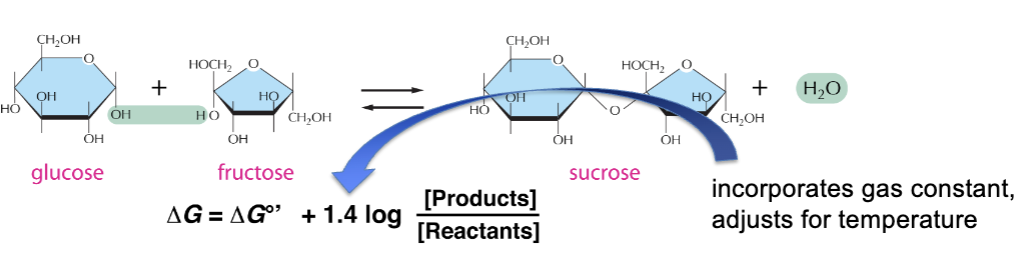
how to make endergonic reactions proceed
ΔG for a reaction is concentration-dependent
– log ([Products] / [Reactants]) is a measure of actual reaction conditions and can be negative
– may offset a positive ΔG°’
Link an exergonic reaction to an endergonic reaction, so that the overall ΔG is negative
(ΔGs for sequential reactions are additive)
ATP (adenosine triphosphate)
main energy carrier in a cell; hydrolyzed to ADP (adenosine diphosphate) with release of energy; one constituent of RNA
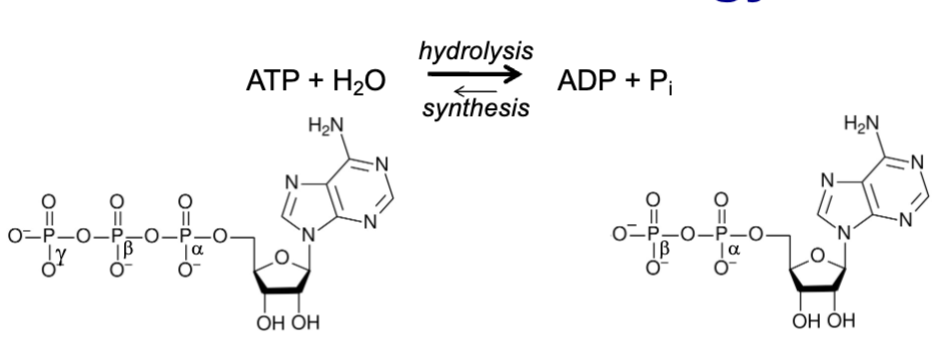
concentration dependent
ΔG is…
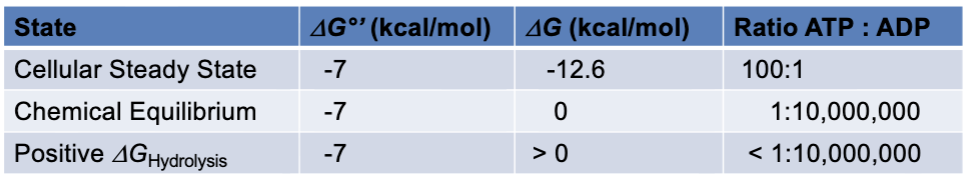
cellular steady state
is not the same as chemical equilibrium; ideal state for a cell to survive and perform the necessary functions
linking reactions
exergonic to endergonic so that overall ΔG is negative
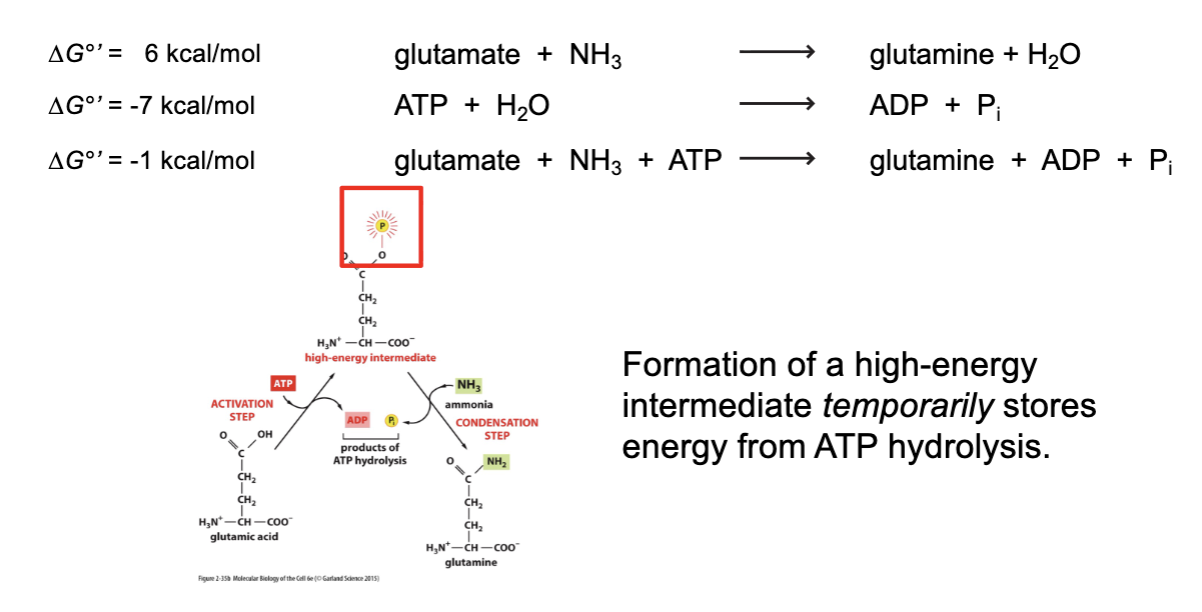
free energy summary
ΔG is the change in free energy for a reaction
only reactions with a negative ΔG can proceed spontaneously
ΔG is highly dependent on molecular concentration inside cells
ΔG for sequential reactions is additive reactions with a positive ΔG°’ can proceed if:
– concentrations of reactants or products are changed, so that ΔG is negative
– coupled to a reaction with negative ΔG
ΔG makes no statement regarding reaction rate.
high energy transition state
Reactions require overcoming a __________ ________ before a reaction can proceed
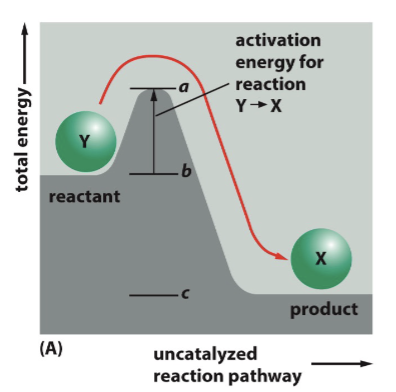
sufficient
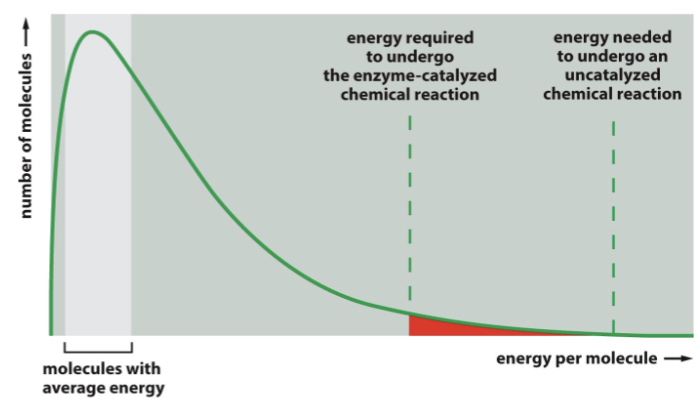
only a small fraction of molecules have _______ energy
enzymes
can lower the energy requirement significantly, thereby increasing the rate of the reaction
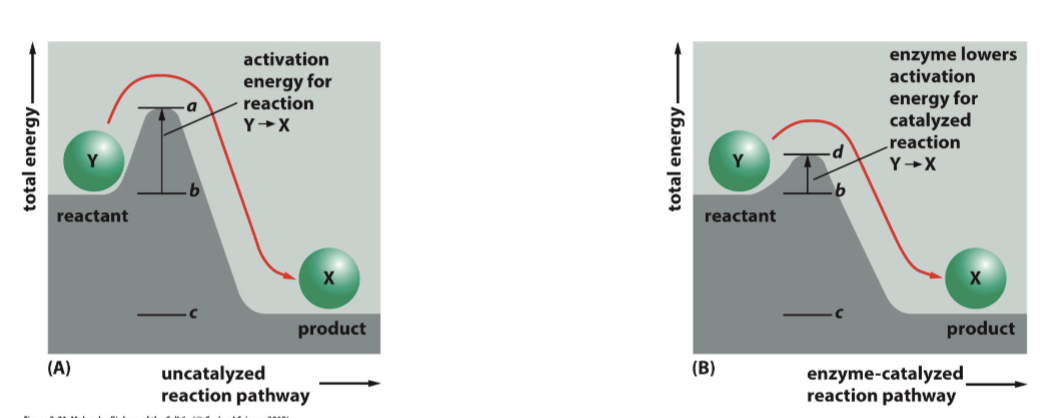
starting and ending energy levels
the number of transition steps and their energy requirements are irrelevant, ΔG is only determined by…
three different ways for enzyme catalysis
A) enzyme binds to two substrate molecules and orients them precisely to encourage a reaction to occur between them
B) binding of substrate to enzyme rearranges electrons in the substrate, creating partial negative and positive charges that favor a reaction
C) enzyme strains the bound substrate molecule, forcing it toward a transition state to favor a reaction
orients
enzyme binds to two substrate molecules and _______ them precisely to encourage a reaction to occur between them
rearranges electrons
binding of substrate to enzyme _______________ in the substrate, creating partial negative and positive charges that favor a reaction
forcing it toward
enzyme strains the bound substrate molecule, ______________ a transition state to favor a reaction
steps in enzyme catalysis
enzyme binds substrate, forming E-S complex – via non-covalent bonds – E and S meet randomly
catalysis happens, E-S complex becomes enzyme-product complex (E-P)
enzyme and product dissociate
Enzymes accelerate rate of reaction,
cannot change K eq!
lysozyme
takes a six-sugar molecule (a hexasaccharide) and cleaves it between sugars 4 and 5.
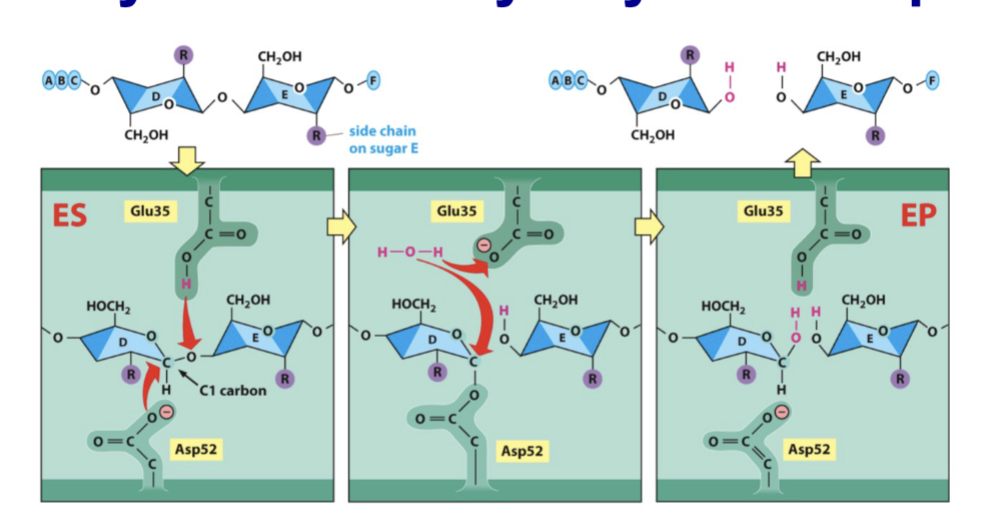
conformation
lowered activation energy because enzyme twists sugars into ___________ resembling activation state
covalent bond
a _________ linking enzyme and substrate forms briefly during catalysis (not true for all enzymes)
reconstitutes
release of product ________ enzyme
noncovalent bonds
enzyme and substrate bind through _____________ (key/lock analogy or induced fit)
higher energy state
some enzymes twist substrate into a _________________, resembling the reaction intermediate
lowers
enzymes ______ the activation energy for the reaction, so more molecules can react
→ enzymatic reaction occurs (faster than unassisted) → enzyme and product dissociate
catalyst
enzymes as _________: must return to their starting state
simultaneous hydrolysis
may meet energy requirement by ____________of high-energy molecules, eg. ATP
protein concentration (levels)
transcription, translation, destruction, and localization
conformational change
activity of protein regulation: regulated transition between active and inactive states
allosteric activators/inhibitors, posttranslational modifications, and complex formation
ways for conformational change to occur
substrate availability and binding to active site
sequestration of substrate; and competitive inhibitors compete with substrate for binding
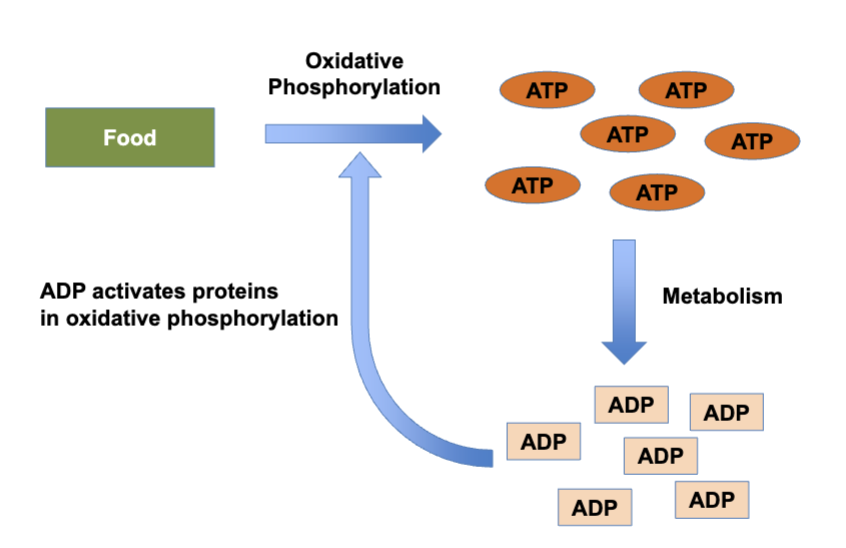
positive feedback
example: as the concentration of ADP increases, the conversion of food molecules into ATP is increased, activating the reaction
arrowhead
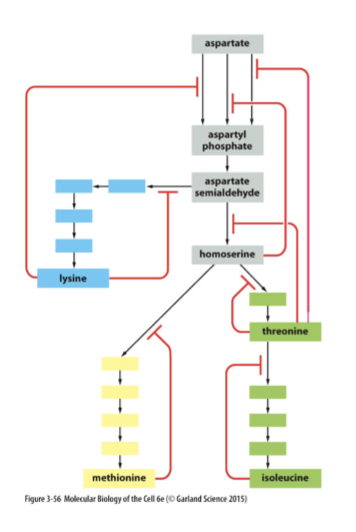
negative feedback
end products of biochemical pathways will inhibit an early step in the pathway (product inhibition), shutting it down
square line
allosteric inhibition
binding usually not at active site, but at a regulatory site
inactivating
binding causes conformational change, _________ enzyme
EF-Tu protein example
switches between active and inactive states; regulated by a small molecule → induces large-scale conformational change
bound to GTP: red domain is -helix, binds tRNA → EF-Tu is active
bound to GDP: red domain is a loop, no tRNA binding → EF-Tu is inactive

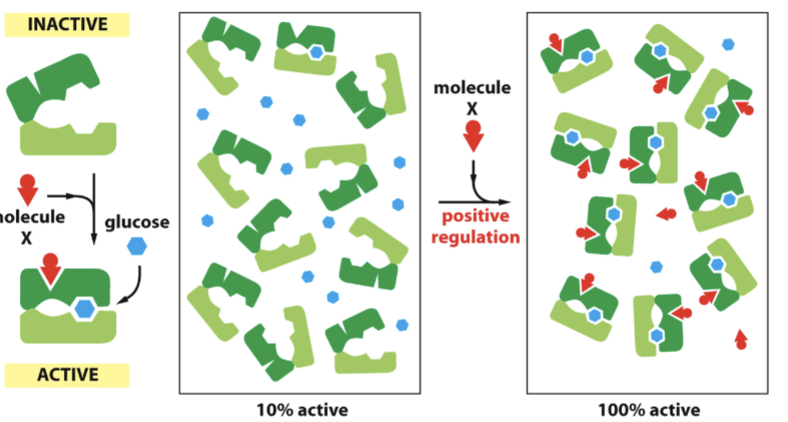
allosteric activation
Binding of activator causes conformational change and the binding of activator and substrate is cooperative.
allosteric inhibition
Binding of inhibitor causes conformational change, which is incompatible with substrate binding.
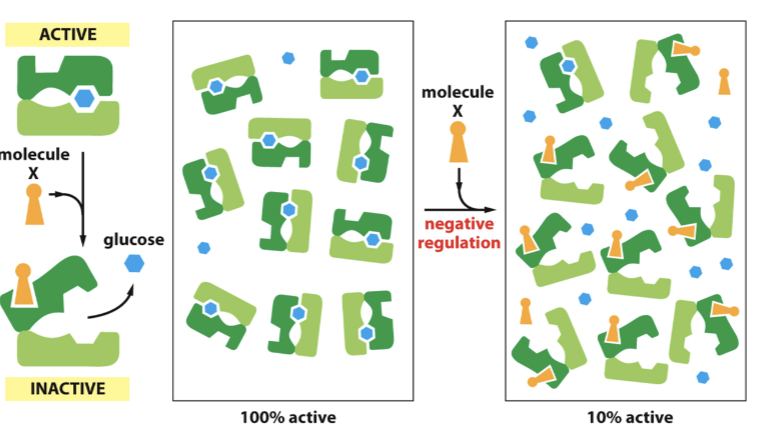
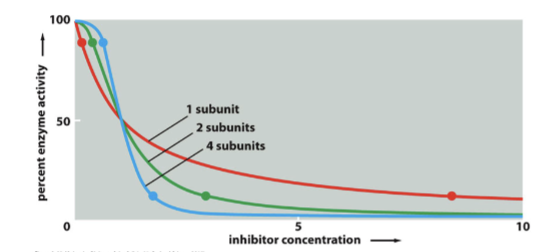
multi-subunit inhibition
addition of first inhibitor is energetically unfavorable → forces change in additional subunits and breaks the symmetry
additional inhibitors bind more easily → restores symmetry and there is cooperative binding
response to inhibitor is much steeper
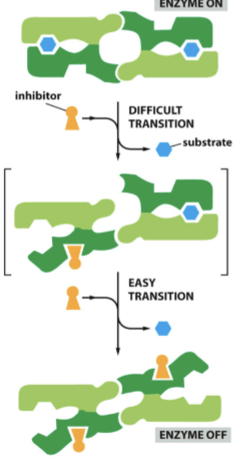
more difficult; entire complex
in multi-subunit inhibition, ___________ to bind first inhibitor, but additional inhibitors bind more easily. Once the first inhibitor binds, the __________ is affected
on/off switch
Inhibition is more rapid and more closely resembles this when there are more and more subunits
competitive inhibition with ATP
ATP-γS is a non-hydrolysable analog of ATP, and its overall shape of molecule is unchanged: expected to bind with similar characteristics as ATP
ATP-γS competes with ATP for binding to the active site → γS strongly inhibits hydrolysis of phosphate ester
allosteric inhibition summary
molecule binds away from the active site, the enzyme conformation is changed, and it affects the rates of substrate binding and processing

competitive inhibition summary
molecule binds at active site directly, enzyme conformation is unchanged, competes with substrate for binding, and the inhibitor is usually chemically inert

competitive or allosteric inhibitor example
LQM is an allosteric inhibitor because it shut down any and all reactivity; increasing the inhibitor meant no product was formed; when there were equal amounts of enzyme and inhibitor, some product was made, and with more inhibitor, regardless of the amount of substrate, zero product was formed.
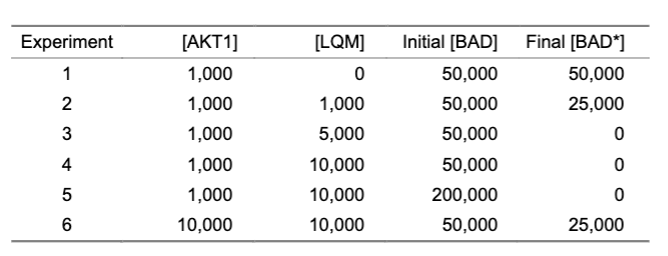
ratios
competitive: increase the amount of substrate = increase in product
allosteric: increase amount of enzyme = decreased inhibition
inhibitor to substrate
ratio that matters for competitive inhibition
overwhelm inhibitor, with some substrate, means product will form
concentration of inhibitor
A factor that matters for allosteric inhibition
With too many binding sites, some enzymes without the inhibitor can process substrate into product;
not dependent on substrate concentration
Posttranslational Modifications
enzymatic addition of groups to polypeptide chains by covalent bonds; they affect: protein shape, ionic charge, protein stability, protein-protein interaction, enzymatic activity, and subcellular localization
common modifications
phosphorylation, methylation, acetylation, prenylation, ubiquitination, sumoylation, glycoslyation
fast and reversible
posttranslational modifications occur because they are …
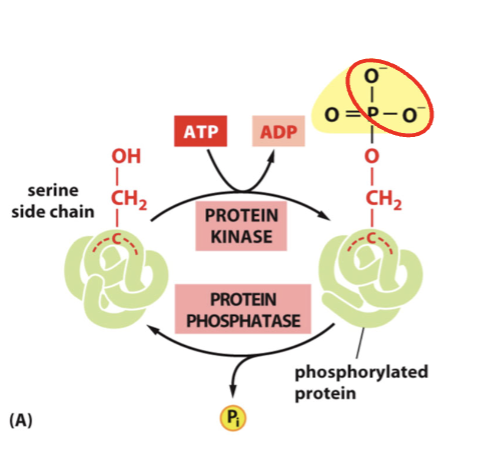
kinase
enzyme that adds phosphate
phosphatase
enzyme that removes phosphate
phosphorylation
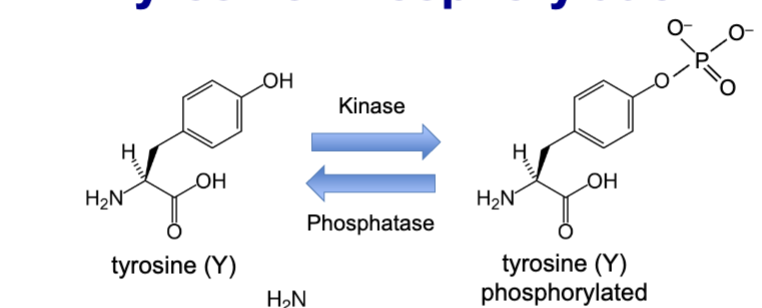
kinase modify specific amino acids: tyrosine, serine, and threonine.
The enzymes kinase and phosphatase add/remove negative charges, and this process is key in cell signaling
tyrosine phosphorylation
kinases transfer the γ-phosphate from ATP
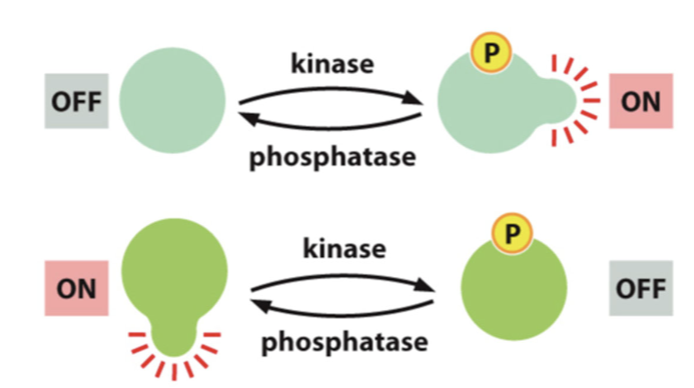
inactive
some proteins become ______ by phosphorylation
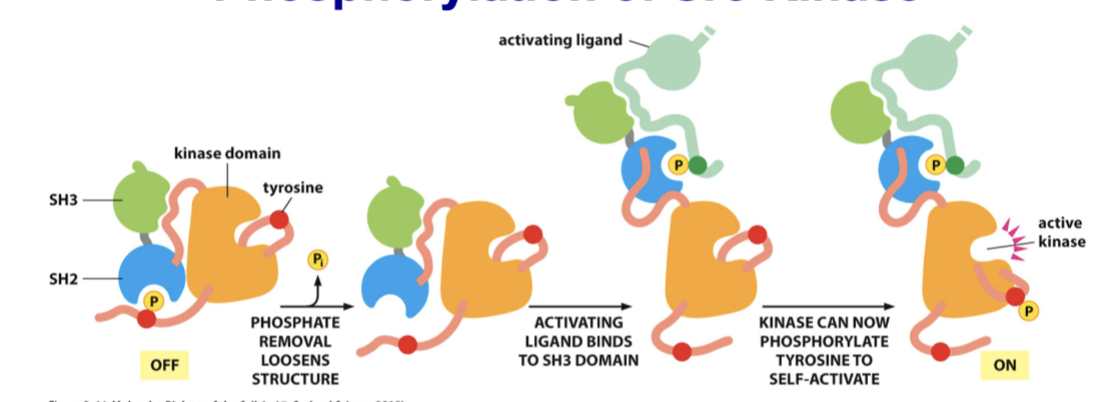
Src kinase
part of a virus that causes cancer in chickens; the viral gene is a copy of the human gene that lacks the c-terminal inhibitory tyrosine
During phosphorylation, the phosphate removal loosens the structure, the activating ligand binds to the SH3 domain, and the enzyme can now phosphorylate tyrosine to self-activate
signal integrators
Proteins can serve as _________
Are certain residues phosphorylated?
Are certain residues unphosphorylated?
Are certain binding partners present?
Are certain binding partners absent?
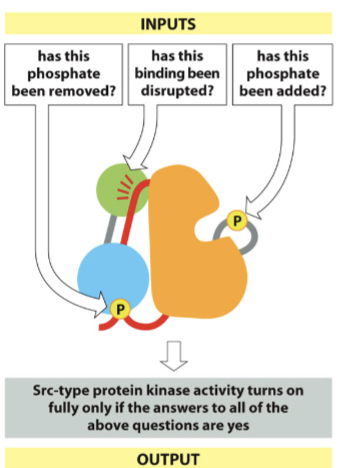
highly regulated
protein activity is _________ through positive and negative feedback
regulation
can be allosteric or competitive
– allosteric regulators bind away from the active site of the protein
– Competitive regulators bind to the active site
Conformational change can regulate protein activity
reversibly
protein activity can be ________ regulated by post-translational modifications
phosphates
added by kinases and removed by phosphatases