Biology Topic 4: Genetic information, variation and relationships between organisms
1/37
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
38 Terms
describe DNA in prokaryotic cells
DNA molecules are short, circular and not associated with proteins (no histones)
describe DNA in eukaryotic cells
DNA molecules are very long, linear and associated with proteins (histones)
describe DNA in the mitochondria and chloroplasts of eukaryotic cells
short, circular and not associated with proteins
what is a gene?
of base sequence of DNA that codes for the amino acid sequence of a polypeptide or a functional RNA (mRNA, tRNA)
where is a gene located?
a gene occupies a locus on a chromosome/DNA molecule
what is a codon/triplet?
sequence of 3 DNA bases
why is the genetic code universal?
the same four bases (A, T, G, C) are used in the DNA of every organism
the same codons encode the same amino acids in every organism
codons in DNA are transcribed into mRNA and translated into amino acids in every organism
why is the genetic code non-overlapping?
there is no overlap between triplet codes - each nucleotide is part of only one codon
[i.e. the codons ATC and CGA are represented like this “ATCCGA” and not “ATCGA” - each nucleotide is only read once]
why is the genetic code degenerate?
multiple codons can code for the same amino acid
[e.g. tyrosine is coded for by TAC and TAT]
does all of the nuclear DNA code for polypeptides?
no - the majority of it doesn’t
there are multiple non-coding bases (introns) between genes
what are the coding regions of DNA called?
exons
what are the non-coding regions of DNA called?
introns
what is a genome?
the complete set of genes in a cell
what is a proteome?
the full range of proteins a cell is able to produce at a given time
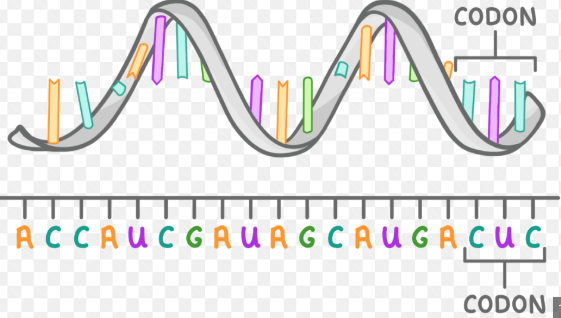
describe the structure of mRNA
single stranded, linear polynucleotide chain
composed of nucleotides (ribose sugar, phosphate group and nitrogenous base)
contains nucleotides adenine, uracil, cytosine and guanine
nucleotide bases are arranged into codons, which specify an amino acid or stop/start signal
nucleotides are linked by phosphodiester bonds
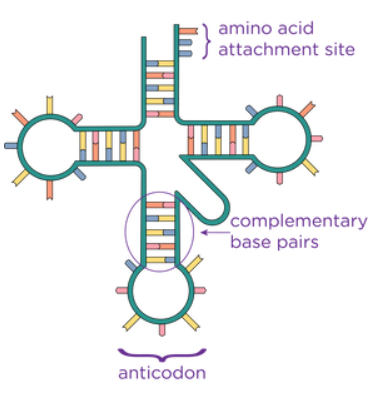
describe the structure of tRNA
single strand folded into a clover shape due to hydrogen bonds between complementary base pairs
one end is attached to an anticodon which corresponds to an mRNA codon, and the other has a binding site for the corresponding amino acid (to the mRNA codon)
what is transcription?
in prokaryotes - production of mRNA from DNA
in eukaryotes - production of pre-mRNA from DNA (this is then spliced to form DNA)
describe the process of transcription (prokaryotes)
RNA polymerase binds to the promoter region of the target gene. it then breaks the hydrogen bonds between complementary base pairs, unwinding the strands. this exposes the nucleotides.
exposed nucleotides on the template strand pair with free RNA nucleotides via complementary base pairing. H bonds are formed between base pairs, and Adenine (DNA) binds with Uracil (RNA)
RNA polymerase binds adjacent nucleotides together via phosphodiester bonds. this forms the mRNA strand. the mRNA strand is a copy of the target gene.
when RNA polymerase reaches a stop codon on the template strand transcription ends and it separates the mRNA strand from the template. the two DNA strands rejoin behind the enzyme. the mRNA strand exits the nucleus via the nuclear pores.
describe the process of transcription (eukaryotes)
RNA polymerase binds to the promoter region of the target gene. it then breaks the hydrogen bonds between complementary base pairs, unwinding the strands. this exposes the nucleotides.
exposed nucleotides on the template strand pair with free RNA nucleotides via complementary base pairing. H bonds are formed between base pairs.
RNA polymerase binds adjacent nucleotides together via phosphodiester bonds. this forms the pre-mRNA strand. the pre-mRNA strand is a copy of the target gene.
when RNA polymerase reaches a stop codon on the template strand transcription ends and it separates the pre-mRNA strand from the template. the two DNA strands rejoin behind the enzyme.
[splicing]
pre-mRNA contains introns and exons
spliceosomes bind to the intronic regions and cut them out of the strand
they then rejoin the exons together, forming an mRNA strand. this then leaves the nucleus via the nuclear pores.
is mRNA complementary to the target gene?
no - the template (non-coding strand) is complementary to the coding strand. as mRNA is complementary to the template strand, it is a copy of the coding strand.
thus it is identical to the target gene
what is translation?
the production of polypeptides from the sequence of codons carried by mRNA
describe the process of translation
mRNA binds to a ribosome in the cytoplasm. 2 codons enter the ribosome at a time.
tRNA binds to the first codon in the ribosome.
tRNA anticodons bind to complementary mRNA codons. on the other side of the molecules, tRNA carries a specific amino acid which correlates to a codon on the mRNA strand.
the amino acid on the tRNA forms a peptide bond with adjacent amino acids on other tRNA molecules. the formation of a peptide bond requires ATP
after the amino acid is joined to the polypeptide, tRNA is released. the ribosome moves along the mRNA strand to the next codons to form the polypeptide chain
what role does the ribosome play in translation?
site of protein synthesis
small subunit binds to the mRNA, large subunit bind binds to the tRNA molecules
what do gene mutations involve?
change in the base sequence of chromosomes
what are the 2 main types of gene/point mutations?
base deletion/insertion (indel) - a nucleotide base is inserted or deleted from the DNA sequence. this results in a frame shift.
base substituition - one nucleotide base in the DNA sequence is replaced by another
what is a frame shift?
where a nucleotide base is inserted/deleted, causing the order of the DNA sequence to shift one base forward or backwards
do all base substitutions produce a different amino acid in the polypeptide chain?
no - the DNA code is degenerate, so multiple codons can code for the same amino acid.
this means that not all mutations cause a significant change
what is a mutagenic agent?
a factor (chemical, biological, or physical agent) that increases the rate of gene mutations in the DNA
what are the two main chromosome mutations?
polyploidy - mutation where an individual has more than 2 sets of chromosomes
non-disjunction - mutation where the chromosomes fail to separate during meiosis, resulting in an individual with one more or fewer chromosomes
what does meiosis produce?
4 genetically different haploid daughter cells
process of meiosis
meiosis I:
prophase I: the chromosomes condense and homologus pairs form bivalents. crossing over occurs between non-sister chromatids.
metaphase I: homologus pairs line up at the centre of the cell. independent assortment occurs, causing genetically different daughter cells.
anaphase I: homologus chromosomes are separated, and chromosomes move to opposite ends of the cell
telophase I: chromosomes reach the poles and nuclear envelopes form. in cytokinesis, the cytoplasm divides. this forms 2 genetically different haploid daughter cells
meiosis II:
prophase II: nuclear envelopes break down
metaphase II: chromosomes line up at the equators of the cells
anaphase II: sister chromatids are separated and move to opposite poles
telophase II: chromosomes reach the poles and nuclear envelopes reform.
cytokinesis occurs, dividing the cytoplasm. this results in 4 genetically different haploid daughter cells.
how does independent assortment result in increased genetic variation?
during metaphase I, homologus chromosomes line up at the equator.
the arrangement of these pairs is random, meaning that the chromosomes which end up in each cell after division is random
how does crossing over result in increased genetic variation?
during prophase I, when the homologus chromosomes form bivalents non-sister chromatids exchange genetic material at the chiasmata
this causes recombination, increasing genetic variation
how can you calculate the number of different combinations of chromosomes after meiosis (wo crossing over)?
using the formula:
no. of combinations = 2n
where n = no. of homologus pairs
how can you calculate the number of different combinations of chromosomes after fertilisation?
(2n)2
what happens to the number of chromosomes during meiosis?
overall, the number of chromosomes halves
meiosis I: 2 haploid cells containing 2 sister chromatids
meiosis II: 4 haploid cells containing a single chromosome copy
how are the outcomes of mitosis and meiosis different?
mitosis produces 2 genetically identical offspring, whilst meiosis produces 4 genetically different offspring
mitosis produces diploid offspring, whilst meiosis produces haploid offspring
how does random fertilisation increase genetic variation?
random fertilisation is the fact that any singular sperm in the multitude can fertilise the egg
this increases genetic variation by producing new allele combinations