Regulatory regions and introns
1/41
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
42 Terms
Promoters 1
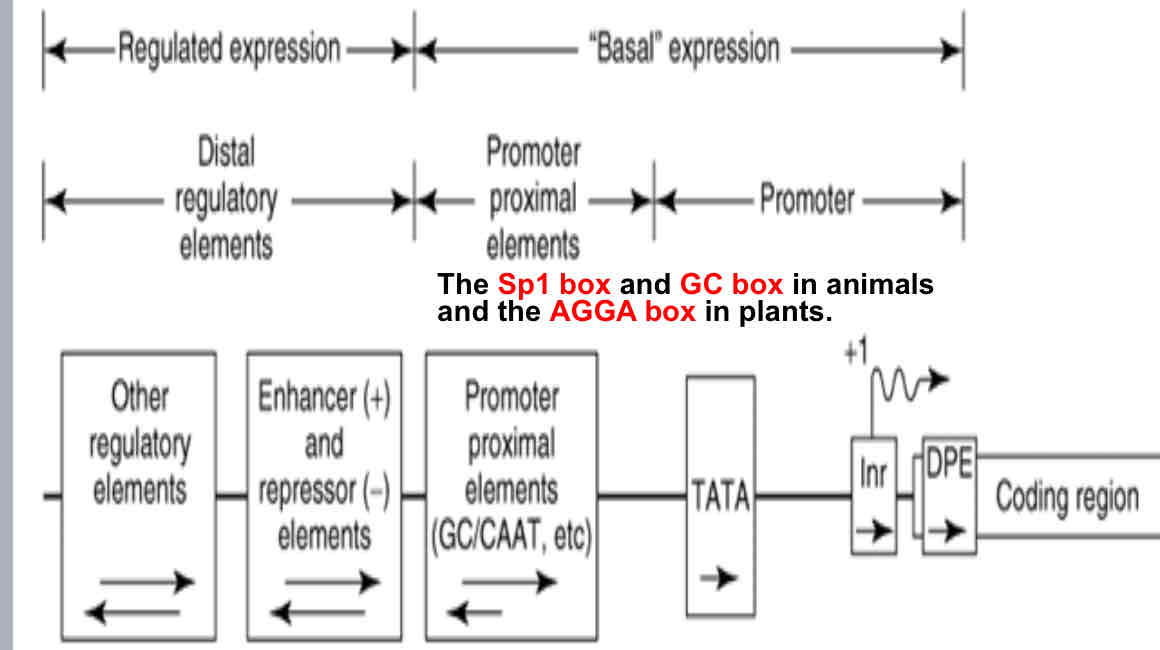
Are regulatory regions of DNA upstream of a gene, which provide a control point for regulated gene transcription. That also determine when and where a genes is expressed
Promoter elements
Core promoter
the minimal portion of the promoter required to properly initiate transcription, located around position -34
Proximal promoter
the proximal sequence upstream of the gene that contains primary regulatory elements, located around position -250
Distal promoter
very ill-defined area which can range from beyond the 5’ end of the gene up to an even into the next gene
Control elements of promoter
Enhancers, silencers and insulators
Eukaryote promoter
Enhancers
Short region of 50-1500 bp of DNA that can bind activator proteins to trigger the transcription of a gene
important role in evolution of species
Developmental biology- development, differentiation and growth of cells/tissues
Invertebrate segmentation
Vertebrate patterning- body axis antero-posterior and left-right
Developmental robustness
Developmental mechanisms- limb development
Promoter silencers
DNA sequences that bind repressor proteins that inhibit expression and act in the opposite way of enhancers
They can reduce or totally inhibit expression
Sequence can be anywhere in the promoter
Binding of repressor proteins may inhibits RNApol complex formation
Can be caused by bending the DNA
Silencer example
NRSF
Neuronal-restrictive silencer factor- produced by rest gene
Repress the transcription of neuronal genes in non-neural cells
When represses REST, NRSF is also inhibited- allowing for the transcription of neuronal genes
Huntingtons disease
Promoter insulators
Genetic boundary elements that block the action of enhancers
sequence that binds proteins can be anywhere
Determine set of genes and enhancer can influence
Stop enhancer working on adjacent genes
Present at boundary of topological association domains (TADs) which divide the chromosomes in- chromosome neighbourhoods
Act by forming loops
Prevent spread of heterochromatin from a silenced gene to an actively transcribed gene
Transposing
Jumping genes
dna sequence that can change position with in the genome
Create mutations and alter size of genome
Transportation often results in a duplication of the transposon
90% maize genome, 50% human genome made up of TEs
Transposon class 1
Reyrotransposons
Copy and paste- from DNA to RNA then, RNA to reverse transcribed DNA
Long terminal repeats
Line 1- transcribed by RNA pol II
SINEs- transcribed by RNA pol III
Transposon class 2
DNA transposon- cut and paste.
Does not involve RNA intermediate
Transpositions are catalysed by several transpositions enzymes
TE- mutagens
Can damage the genome and produce diseases like haemophilia A and B
Severe combined immunodeficiency, porphyria, predispostistion to cancer and Duchenne muscular dystrophy
Epigenetic mechanisms - 1
Variations that are caused by external or environmental factors
Do not later DNA sequence
Alter gene expression levels
Covalent modification
Epigenetic mechanism
DNA methylation and hydroxymethylation
Protein acetylation, methylation, phosphorylation, ubiquitination and sumoylation
MicroRNAs
Epigenetic mechanisms
Non-coding RNAs of 17-25 nt long
MRNA methylation
Epigenetic mechanism
MRNA plays a critical role in human energy homeostatsis
SRNAs
50-250 nucleotides
Virulence gene pathogens
Prions
Epigenetic mechanism
Infectious forms of proteins
Structural inheritance
Epigenetic mechanism
Seems existing structures act as templates for new structure
Nucleosome positioning
Epigenetic mechanism
Nucleosome position is not random, and determine the accessibility of DNA to regulate proteins. This determines differences in gene expression and cell differentiation
Introns
Non-coding sequences in the gene that do not appear in their mature mRNA and they interrupt the coding sequence
Intron group 1
Found in nuclear, mitochondrial and chloroplast genes coding for rRNAs, mRNAs an tRNAs
Introns group 2
Found in primary transcripts of mitochondrial chloroplast mRNA in fungi, algae and plants
A lariat is formed before splicing occurs
Introns group 3
Largest class of introns
Found in nuclear mRNA primary transcripts
Lariat is or med and a RNA protein complex of nuclea ribonucleoproteins is required
RNA is called small nuclear RNA
Introns group 5
Found in certain tRNAs
Splicing endonucleoases cleaves the phosphodiester bond at both ends of the intron
2 exons are joined by a similar mechanism to DNA ligament reaction
Intron structure
Sequence of the intron always starts with GU and ends with GU
Sequences next to GU/AG and also another in the middle are all similar in each intron
Branch point is a iced distance from 3’ end
Splicing of introns
Spliceosome cuts the intro and rejoins the exon of the mRNA
Consists of several proteins
Intron splicing group III
Small nuclear RNAs bind to donor sequence, forming spliceosome complex
2 transesterfication reactions exercise the intron- forming a loop structure (lariat)
The exons are then lighted to form mature mRNA
Intron splicing extra
The spliceosome cuts 5’ boundary
The 5’ end of the intro is joined to the branch point
The spliceosome cuts the 3’ boundary
The exons are joined and the lariat is released
The snRNAs are released
Lariats
Introns exercised into a loop, by 2 transesterification reactions
Why do genes have introns
To regulate gene expression
control elements such as enhancers found with in introns
Introns allow for differential splicing
Evolutionary relics
Alternative splicing
Process that allows the DNA to code for more than one protein by varying the exons present in a mRNA
40% of humans
Can encode many related proteins with 1 gene
Exactly which intron boundaries are spliced is decided by protein factors that bind to the intron boundries and either help or hinder cutting by the spliceosome
Some genes can be spliced in different ways
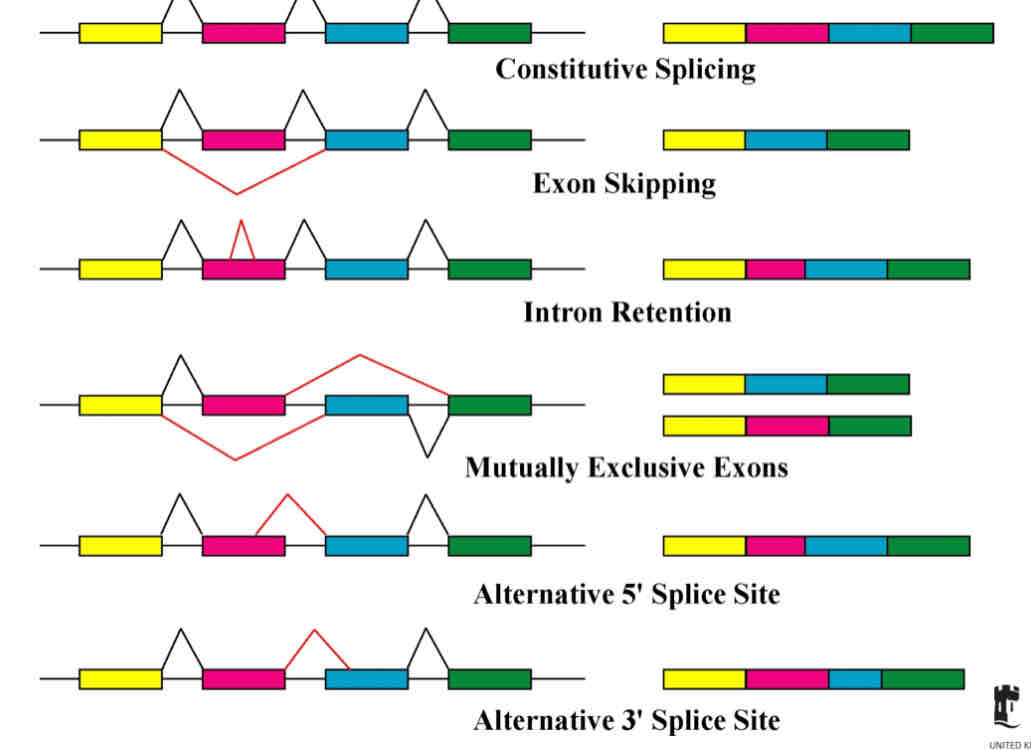
Intron retention fate
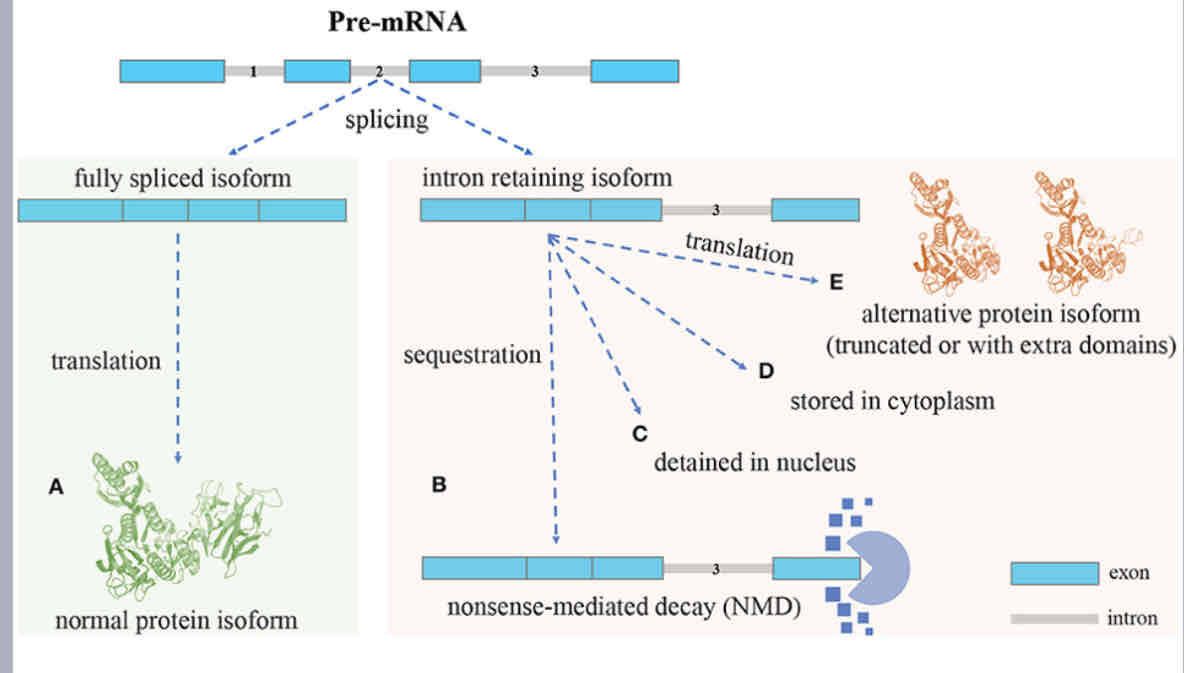
Impact of alternate RNA splicing
5’UTR- altered translation, Change ion protein expression
Protein coding sequence- Altered protein structure and function, change in protein is oformratios
3’UTR- mRNA instability, change in protein expression levels
Alternative splicing 3
The same pre-mRNA can make many different mRA transcripts and may different proteins
Exon order is always preserved
Introns are always discarded
Protein domains
Regions within proteins that fold up in a semi-independent wat and have independent function
Unfolded protein → partially folded protein → fully folded protein
Evolution of introns
Archaebacteria (introns uncommon)
eubacteria (introns very rare)
Eukaryotes (almost every gene has introns)
Intron early theory
Supports idea that introns and RNA splicing were relics of the RNA world
Both eukaryotes and prokaryotes had introns in the beginning
Prokaryotes eliminated them to be more efficient, while eukaryotes kept them to have more genetic plasticity
Introns late theory
Supports that prokaryotic genes are similar to the ancestral genes and introns were inserted later in the gene of eukaryotes
Exon shuffling
Molecular mechanisms to form new genes
2 or more exons from different genes can be brought together ectopically, or the same exon can be duplicated, to create a new exon-intron structure
Exon shuffling mechanism
Transposon mediated exon shuffling.
Crossover during sexual recombination of parental genomes.
Illegitimate recombination.
Question
1- what are promoter elements
2-what are enhancers, silencers and insulator transposon
3- describe the Epigenetic mechanisms
4- what is the intron structure
5-