Communication in unicellular organisms LECTURE 3
1/36
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
37 Terms
Two key levels of communication identified in unicellular organisms
Communication with other members of cell population
Sensing and response to environmental stimuli
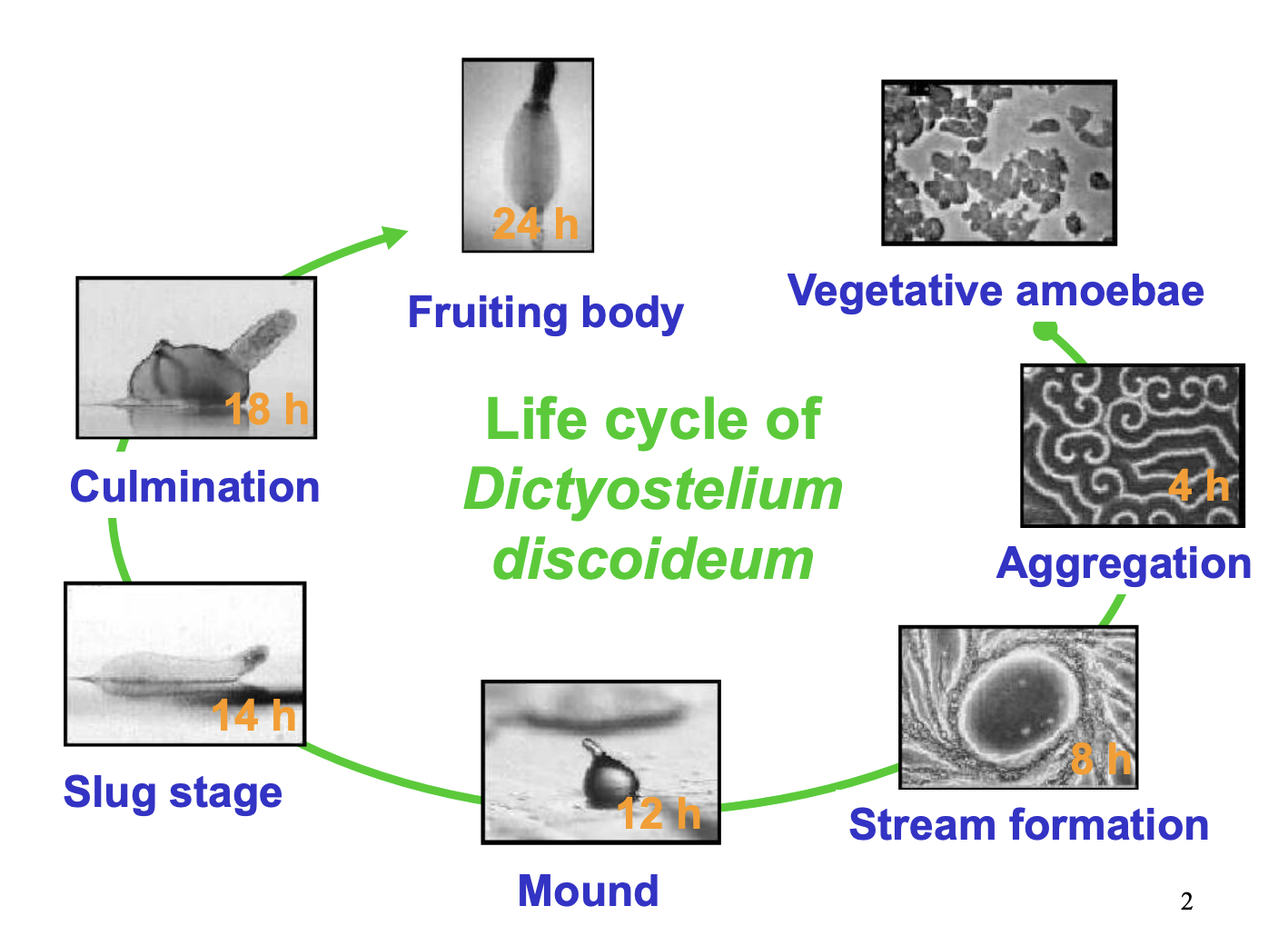
Life cycle of slime mould
How is aggregation initiated
founder cell begins to emit a pulse of cAMP
Attracts neighbouring cells
Neighbouring cells emit own pulse of cyclic AMP→ acts as relay signal
Result: waves on cAMP radiating from aggregation centre every few mins→ 2cm/hour
Cells respond by chemotaxis towards founder cell
How is signal realy and cehmotaxis initiated?
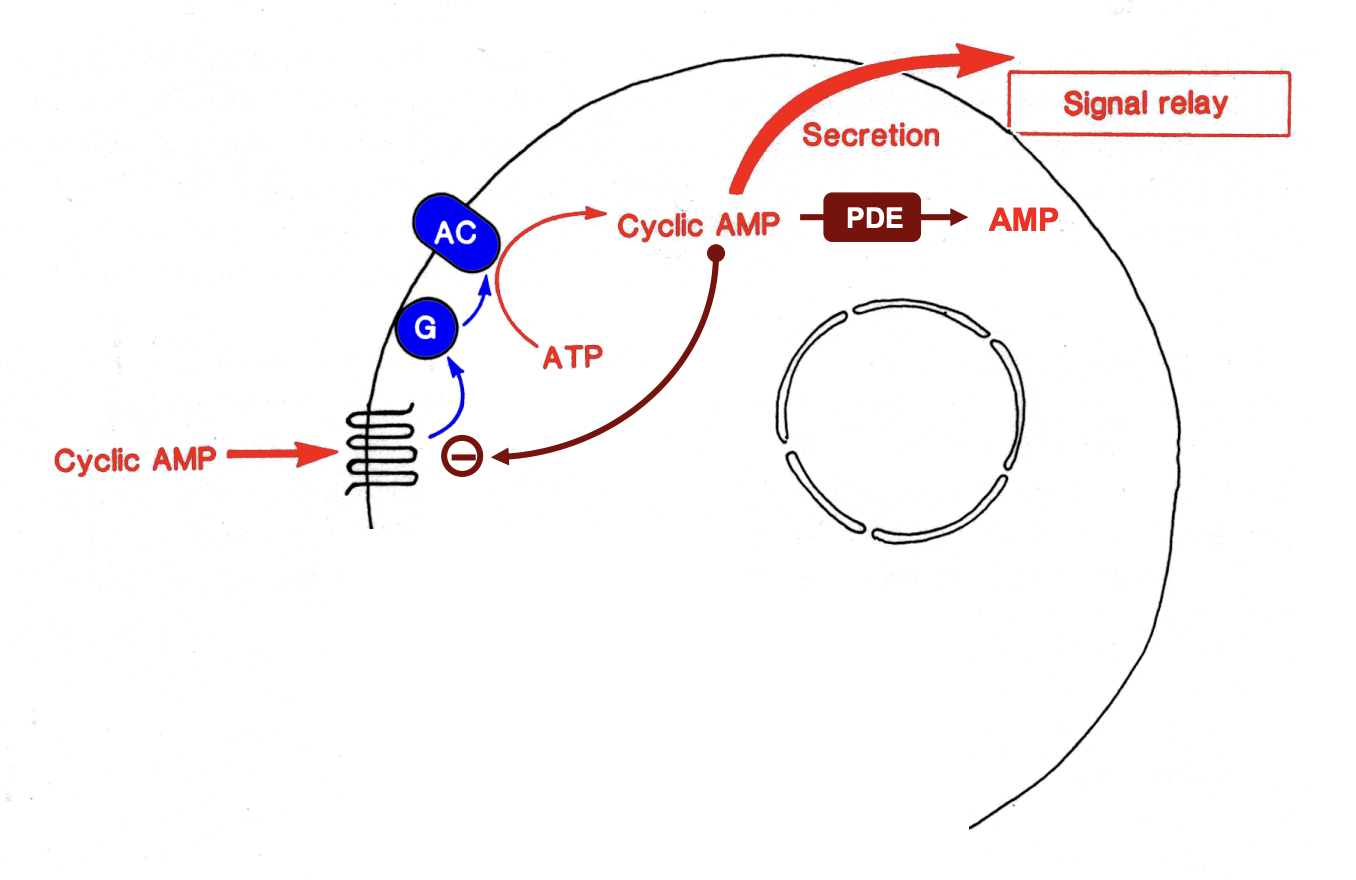
CONTEXT: starvation→ stress→ releases cAMP
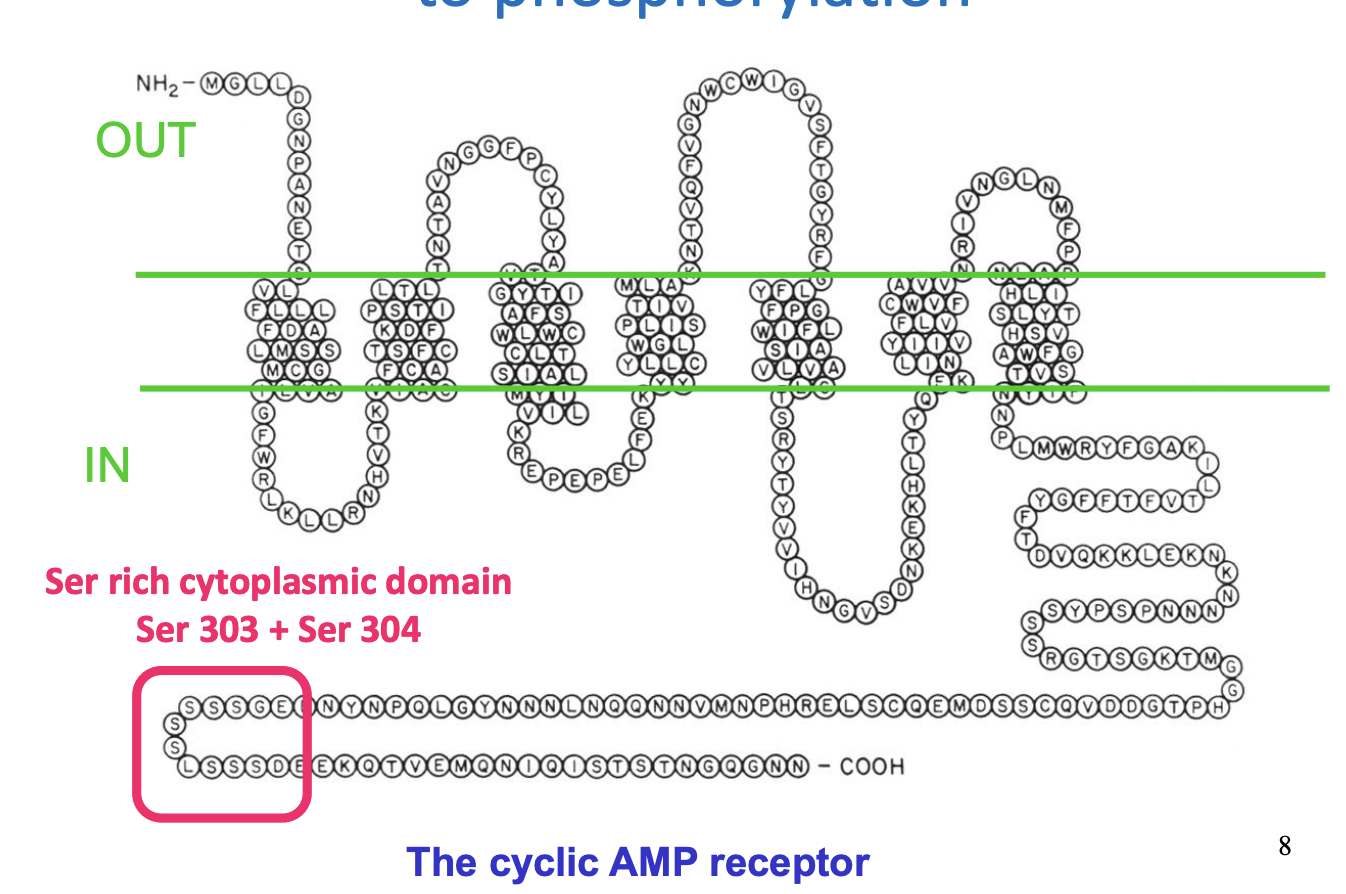
cyclic AMP receptor
→ has 7 helical transmembrane segments
one of the family of G-protein activating receptors
What does cyclic AMP receptor do?
Activate a number of signalling pathways
adenylate cyclase dependent cyclic AMP pathway
→ relay system
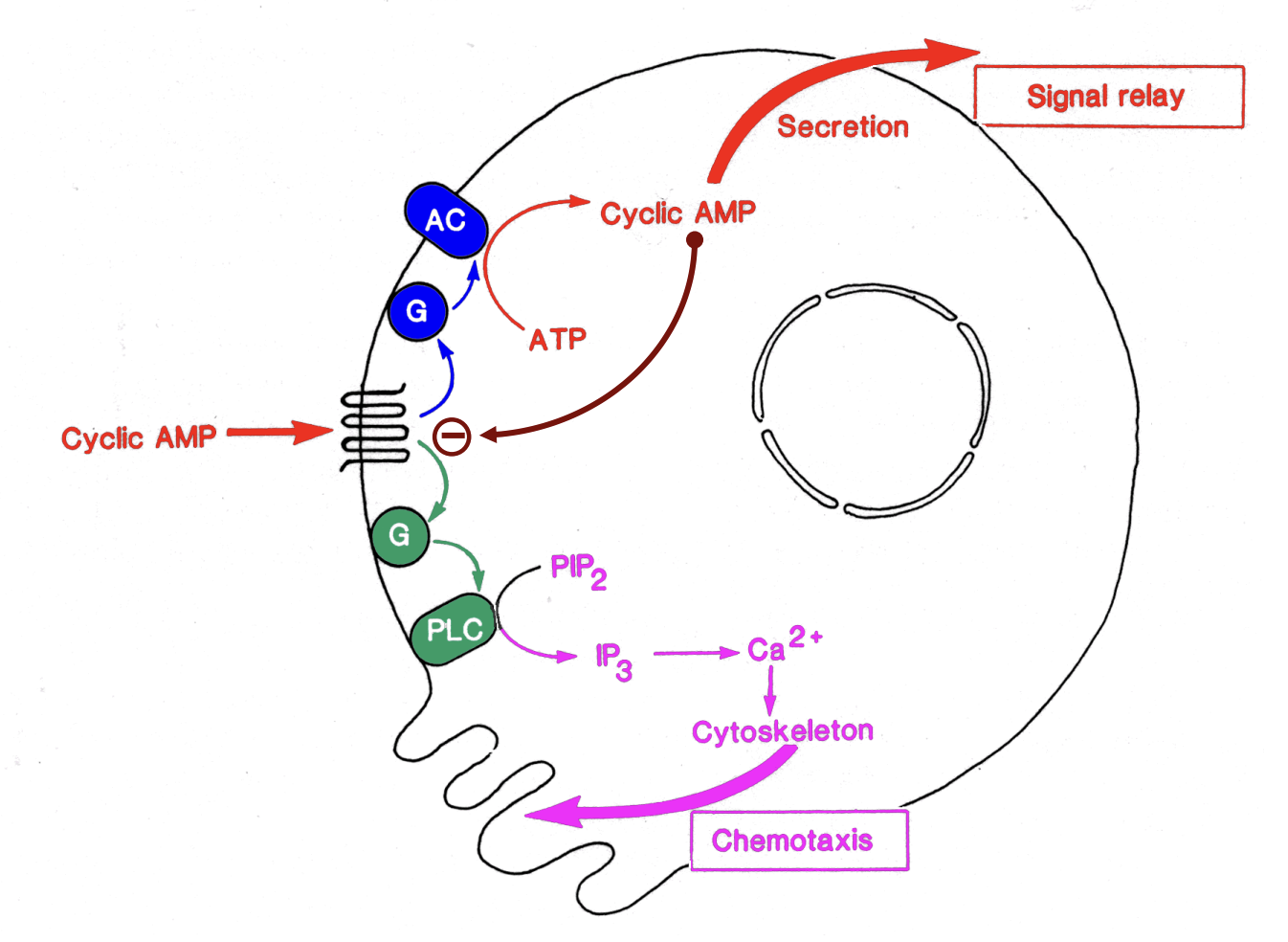
phospholipase C dependent phosphoinositide pathway
→ Triggers local cytoskeletal rearrangements →chemotaxis
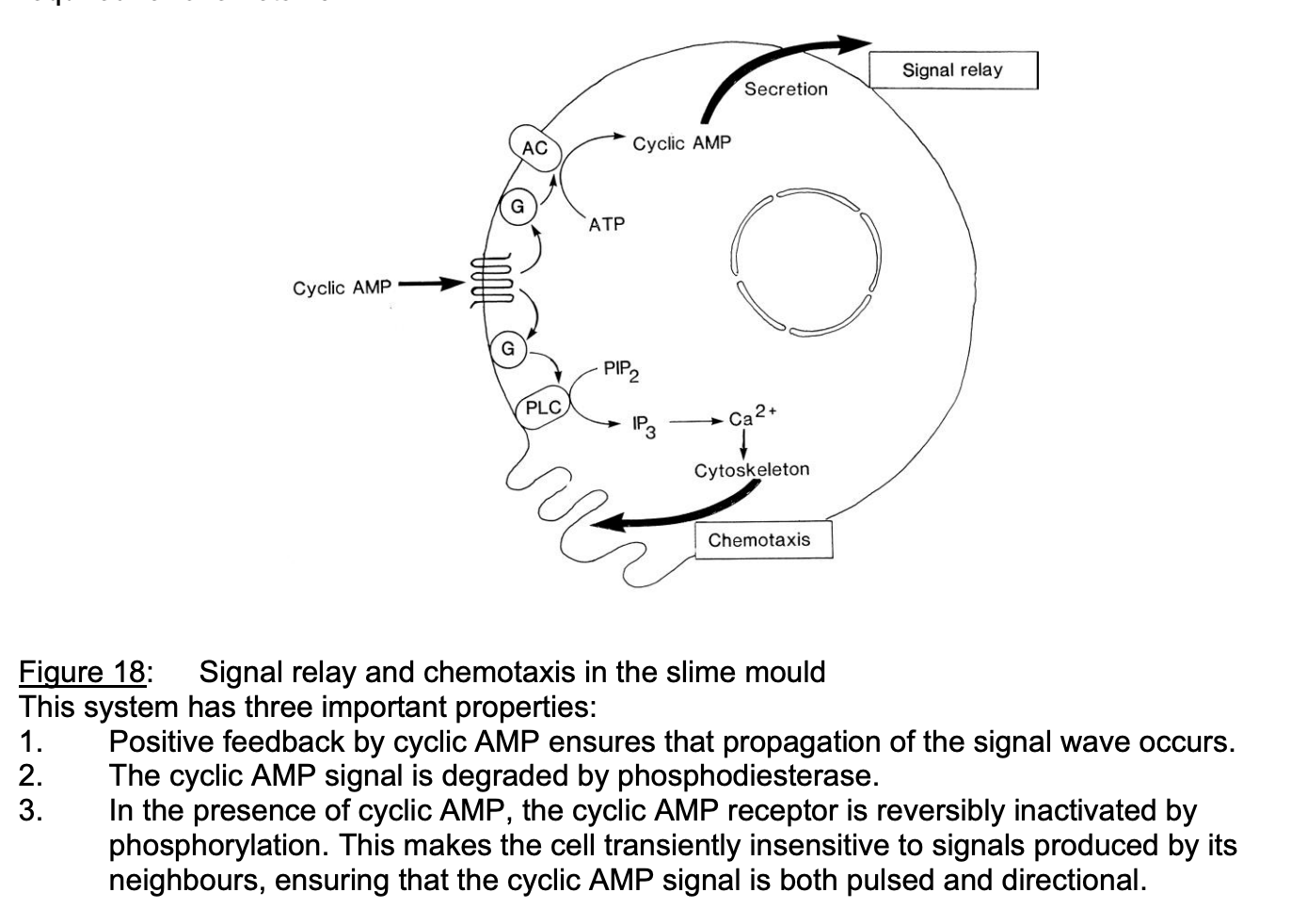
Three properties of signal relay and chemotaxis
Positive feedback by cyclic AMP→ ensures propagation of signal wave occurs
Cyclic AMP signal is degraded by phosphodiesterase
In presence of cAMP, cyclic AMP receptor is reversibly inactivated by phosphorylation
makes cell transiently insensitive to signals produced by neighbours
ensures: cAMP signal is pulsed and directional
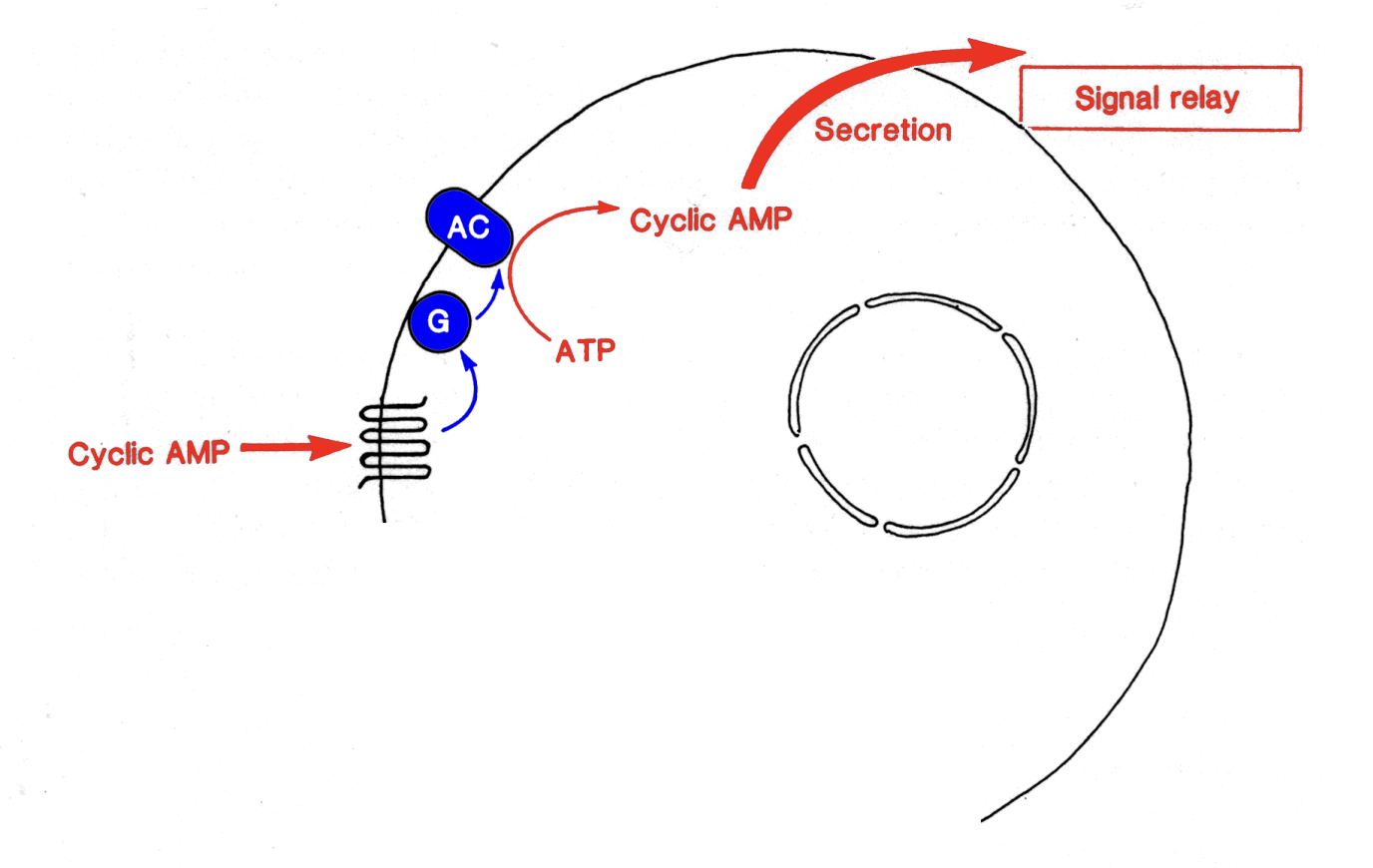
cAMP chemotaxis in slime mould 1
cAMP from outside bind to receptor
comformational change component 3→ G protein→ adeylate cylcase ATP→ cAMP
More cAMP packed into vesciles→ released elsewhere
OVERALL: sends to other cells→ positive feedback
cAMP chemotaxis in slime mould 2 Creating oscillating dynamic
High cAMP conc in the cell
→ causes desensitissation of the cAMP recetepor
BUT ALSO
cAMP→ degraded into AMP
Resensitise of receptor
→ Causes oscillations of desensitisation and resensitisation
These sensitiation oscillations happen due to phosphylation of receptor adatpatation
Phosphylation→ can bind cAMP
Desphosphylation→ cannot bind cAMP
cAMP chemotaxis in slime mould 3 Localaised extension
Only are points in the cell with highest cAMP still there
activates Gq recetpros→ PLC→ PIP2→ IP3 +DAG
IP3→ Ca2+ cytoskeleton
Chemotaxis
elongation of the pseodpods
This process has been tested with mathematical models
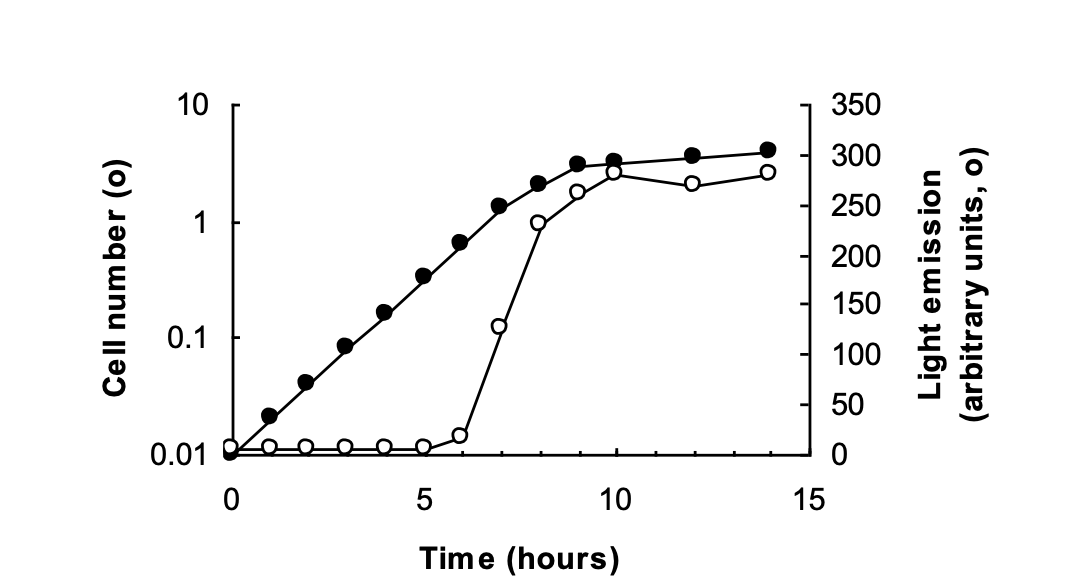
How do bacteria determine population size
Quorum sensing
Why do bacterium Vibrio fisheri use quorum sensing
emission of light is growth phase dependent
only induced when bacterial population raeches a critical cell density
→ dependent on expression of luxA and luxB enoding enzyme luciferase
→ Quorum sensing regulated by Luxland LuxR
OVERALL: only activated when it actually become beneficial
if not enough of them→ light to dim to have any value
How is the quorum sensing signal produced?
OHHL synthesised by Luxl operon
freely diffusible across the cell membrane into external growth medium
Luxl constitutively expressed during exponential growth
concentration of OHHL is directly dependent on the bacterial cell density
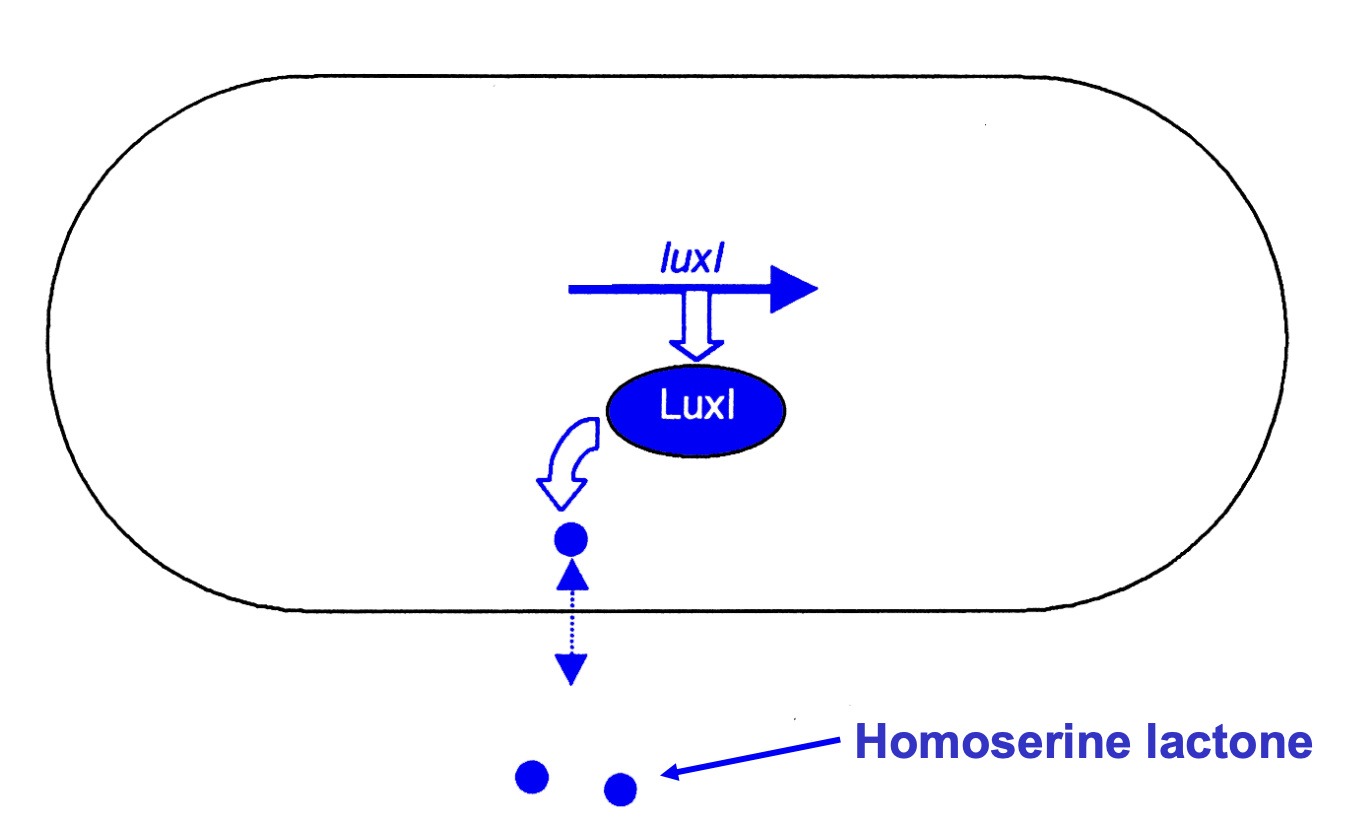
How is OHHL conentration regulated?
Sensed by transcriptional regulator LuxR
OHHL leaves and moves back into the cells continuously
When OHHL reaches critical concentration for luxR binding (must mean there are enough cells around
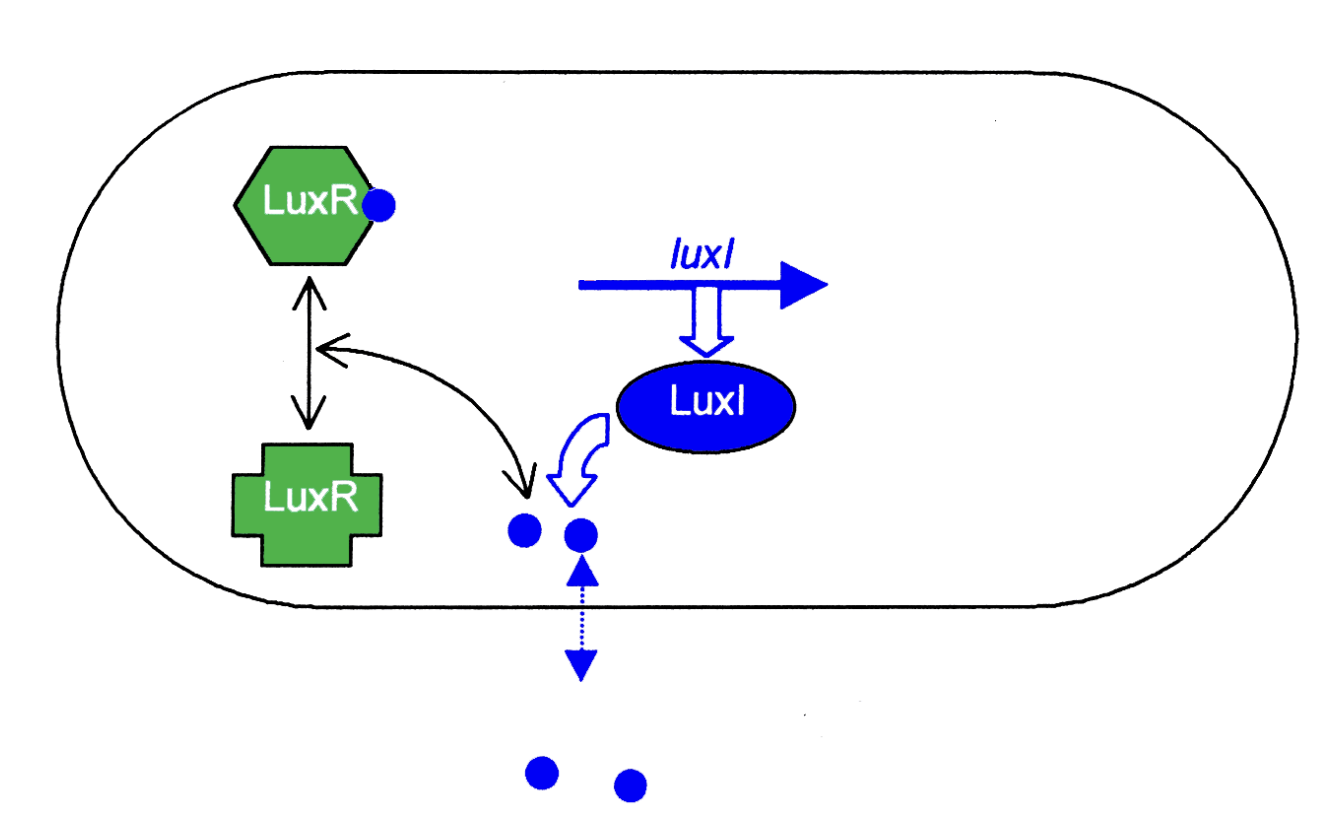
What does LucR induce
LuxR induces 2 sets of genes:
luxl → more OHHL synthesis (positive feedback loop)
ensures all cells are induced at the same time
second cell density dependent target genes induced→ response
lucuciferase
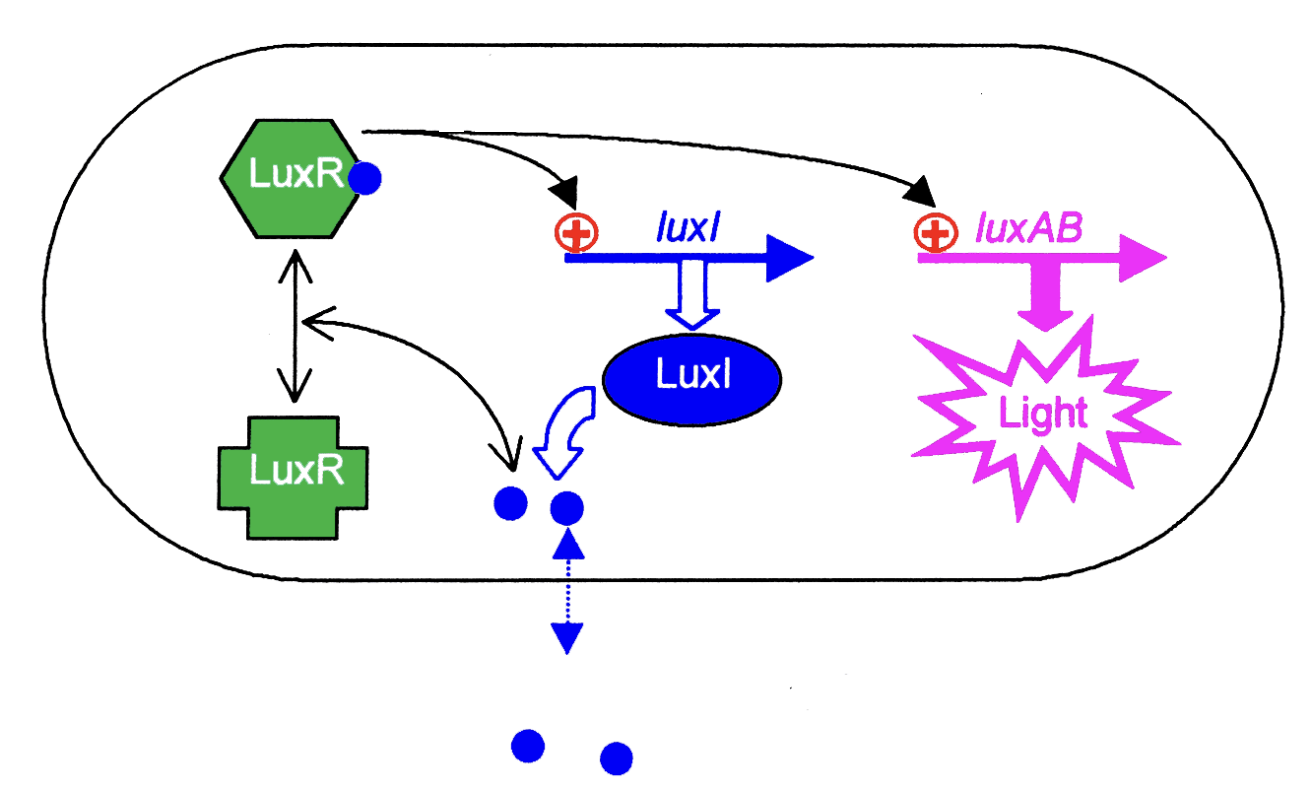
What is the response in this case?
bioluminescence due to
expression of luxA and luxB
Other examples of function regulated by this process?
antibiotic production
extracellular enzyme secretion
→ by pathogenetic bacteria
What does chemotaxis help bacteria do?
Respond to chemical gradients
What do bacteria use to do this?
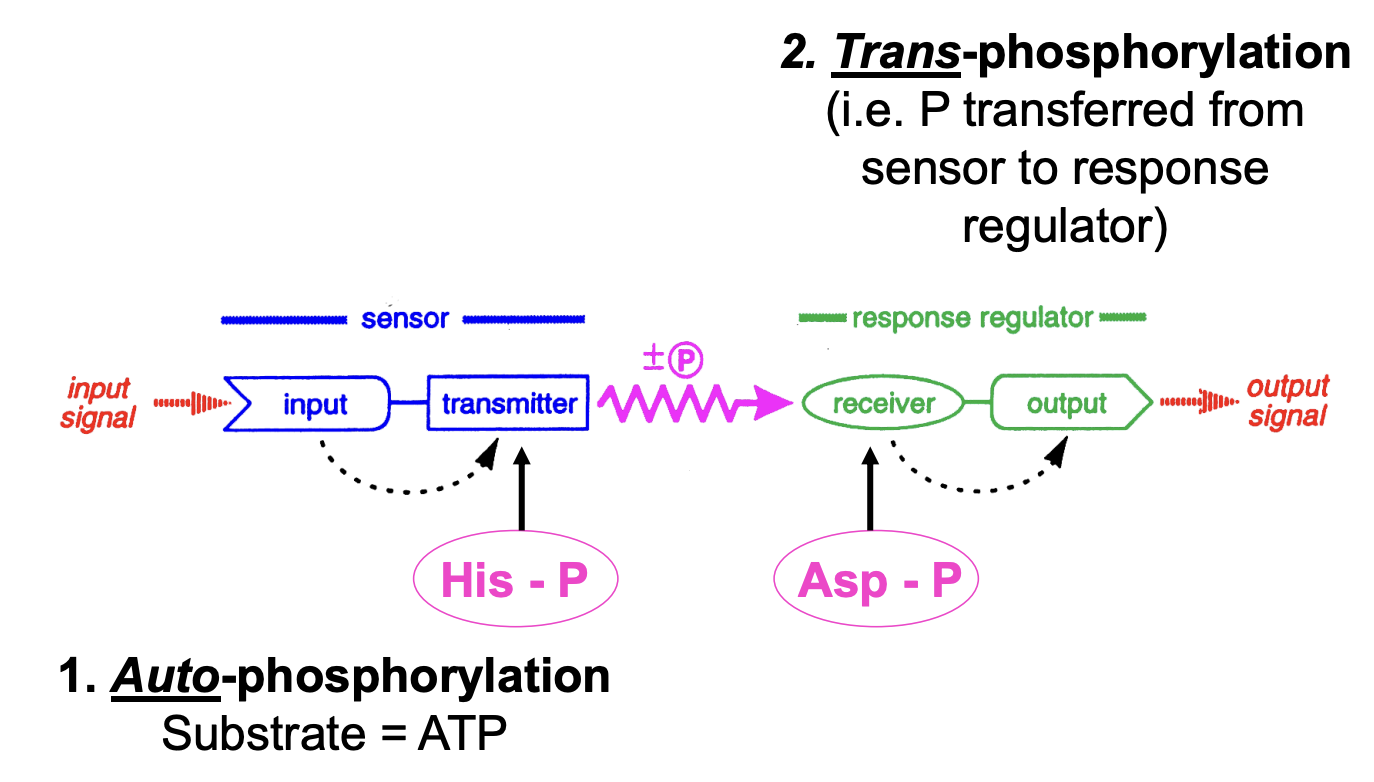
Two component sensor regulator systems:
highly conserved protein pairs
What do these comprise?
Environmental sensor
gets phosphylayed at a conserved histidine residue when a signal is received by the input domain
Auto-phosphylation
Response regulator
Senosr P is psated to receiver (trans-phosphylation)
phosphorylated at conserved asparatate residue by cognate sensor→ activate output domain
phosphate physically transferred from sensor protein to response regulator→ ouput
This then act at either:
DNA level → alter transcription
Protein level→ regulate protein function
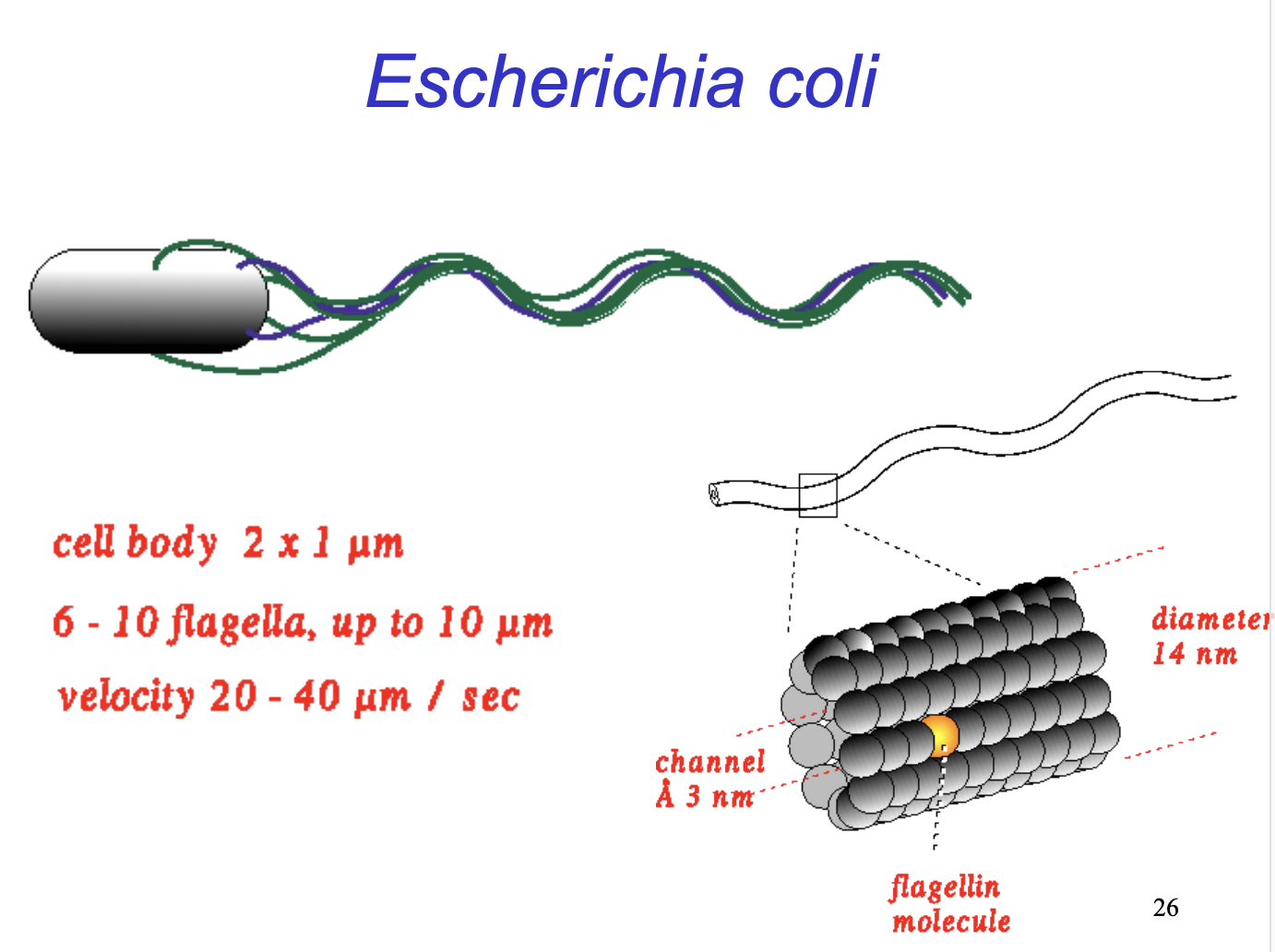
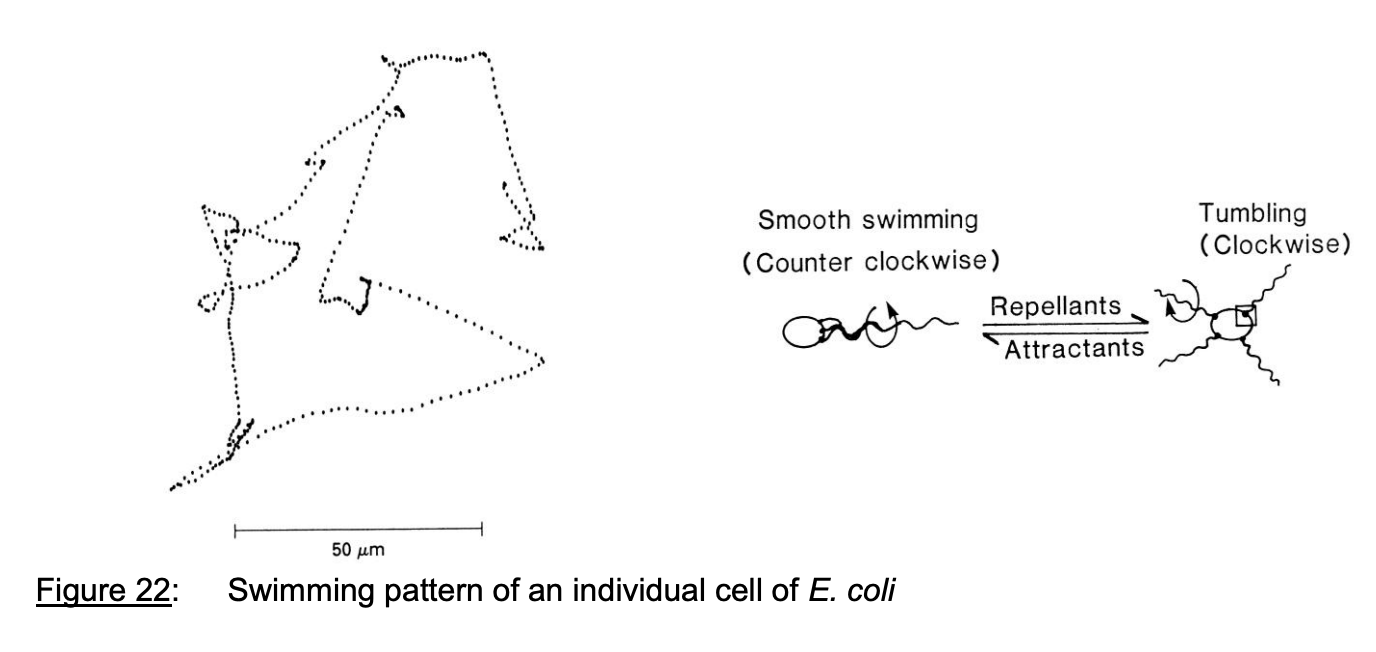
How does chemotaxis move E.coli
10-50 umsec^-1
Towards attractants
Away from repellents
How does the bacterium move?
Rotation of 6-10 flagella
helical protein filametns made up of flagellin
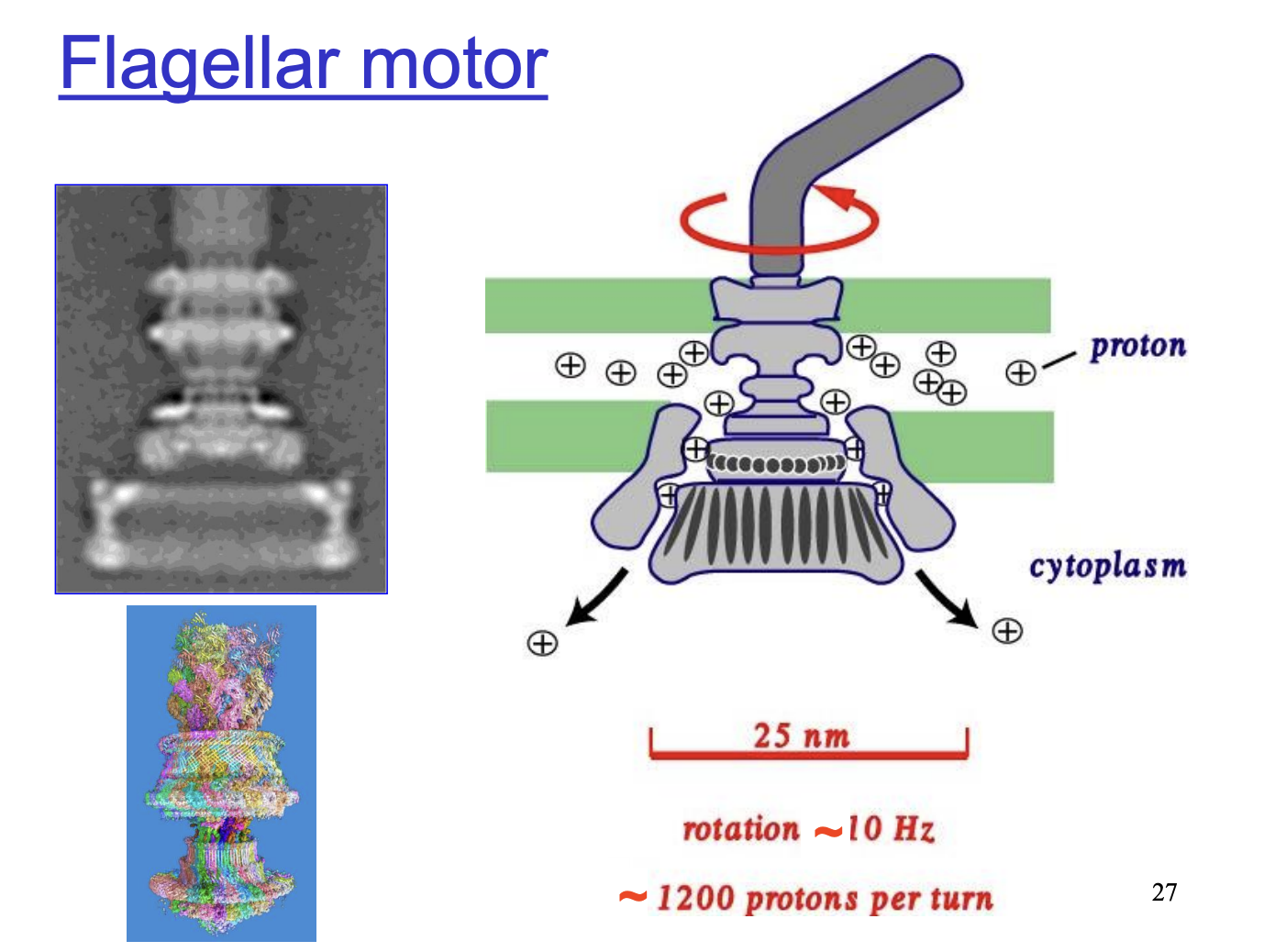
Flagellar motor facts
located in cell membrane
driven by proton motive force
Reversible
Rotates at 10 Hz, requiring 1200 protons per turn
What are the sensor components in this case?
Methyl-accepting chemotaxis proteins (MSP)
theres are at the other end of the flagella
THEREFORE: must be some kind of signalling pathway to get to flagella movement
Flagella rotate counter and clockwise
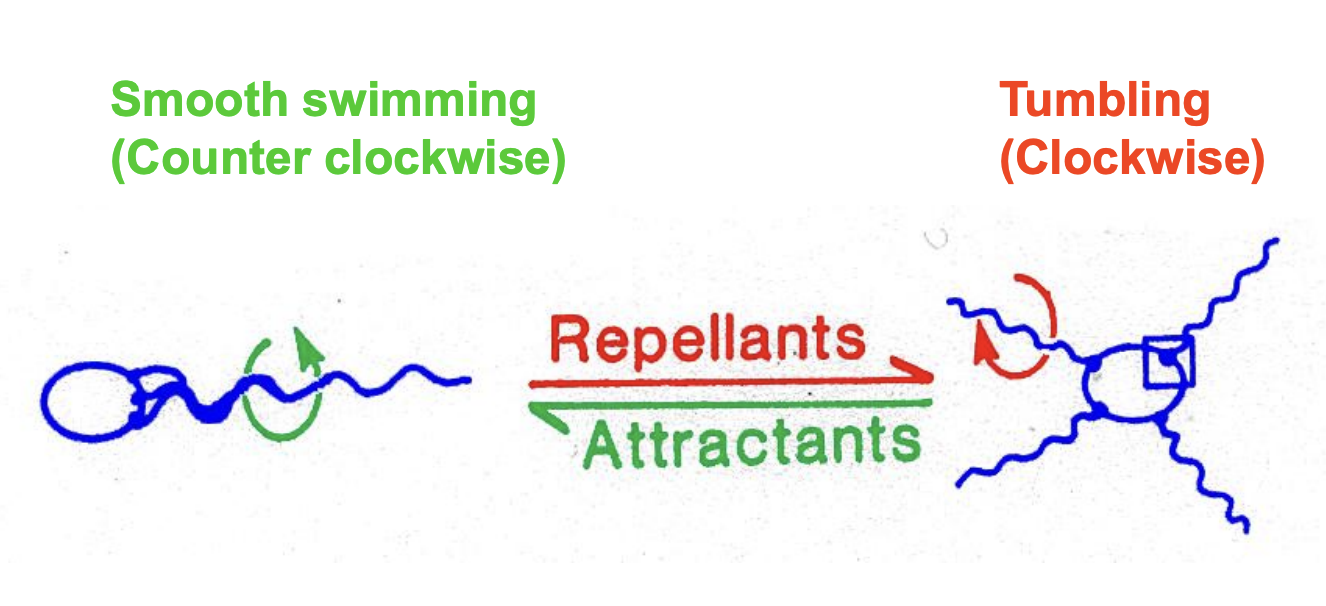
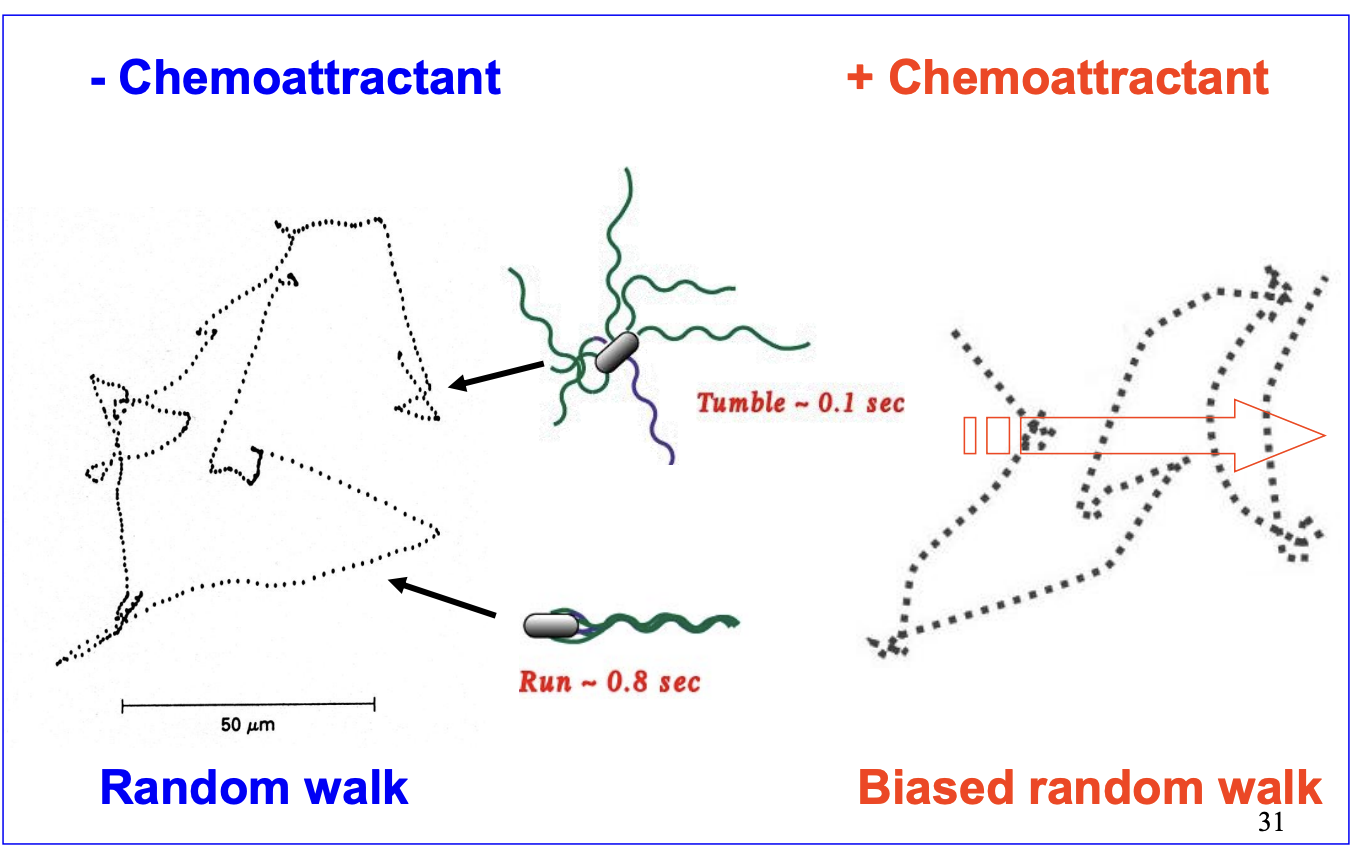
How does chemotaxis work?
Biased random walk
cell swim in relatively straight lines → runs
rotating flagella in counter-clockwise direction for 1 sec
Then reverse flagellar motar for 0.1 sec (tumble)
New run in random direction
Length of run is biased→ runs are longer in favourable and shorter in unfavourable directions
How are changes in concentration of attractant/repellents detected?
Temporally
NOT a spatial sensing mechanism
Biased random walk when there is a chemattractant→ longer runs, shorter tumbles
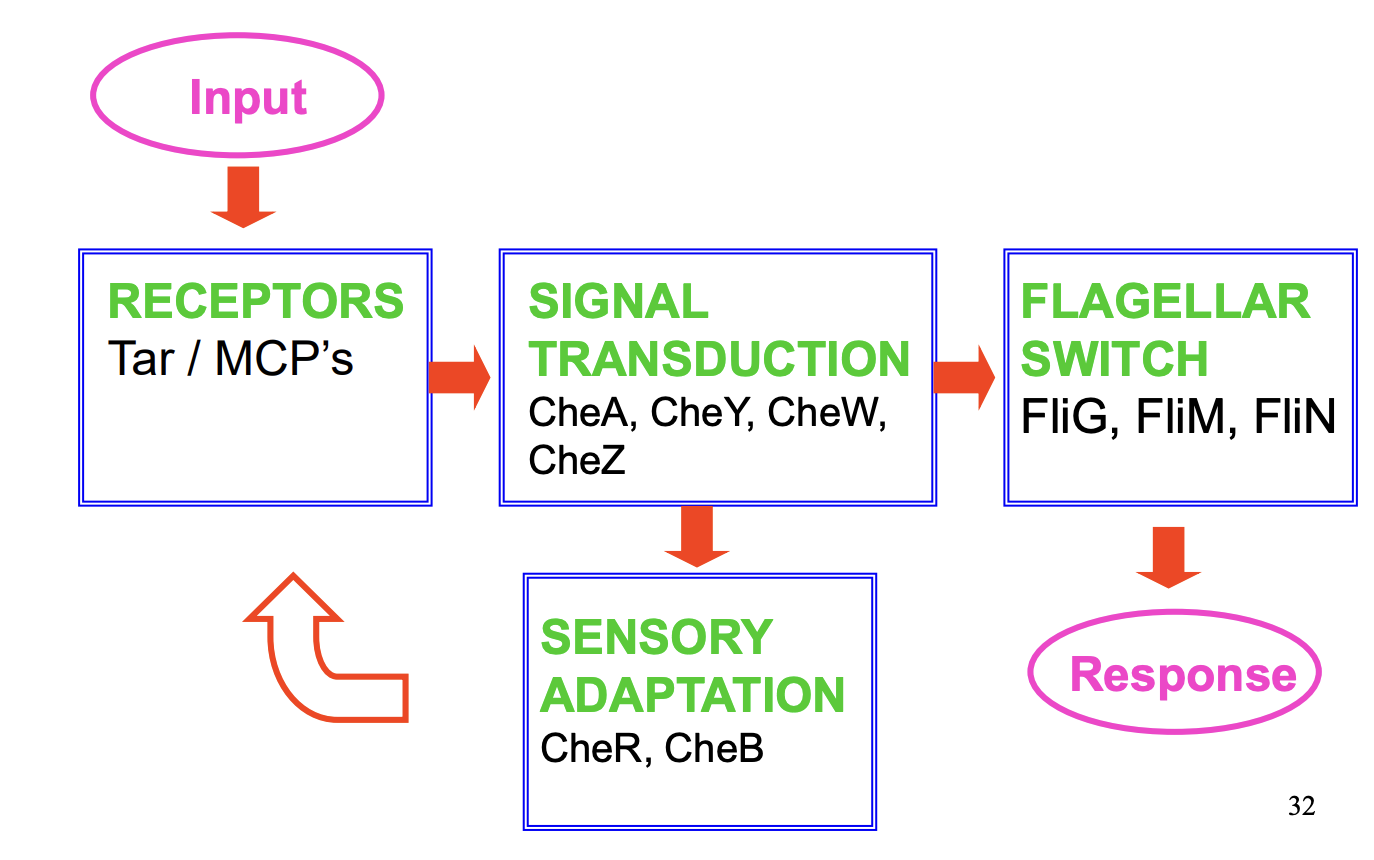
4 sets of proteins which help carry chemotaxis out
Membrane bound signal transducer→ E.Coli has 5 receptor protein classes (MCPs) proteins (methyl accepting chemotaxis proteins)
mediate response to sepcific chemoattractants
e.g Tar→ aspartate and maltose
- > SENSOR
Cytoplasmic signal transduction
CheA (histidine kinase), CheY (response regulator), CheW (adapter protein) and CheZ (an accelerator of CheY dephosphylation)
→ NB. ALSO THE SENSOR BUT A SEPARATE PROTEIN (unlike before)
Flagellar switch
FliG, FliM and FliN
determine direction of flagellar rotation
Adaptation
CheR (methyl transferase)
CheB (response regulator which acts as a methylesterase)
→ controls level of methylation of the chemotaxis receptor proteins
These help with the temporal memory of the concentration gradient thing
How flagella proteins organised
Slightly different type of 2 component sensor
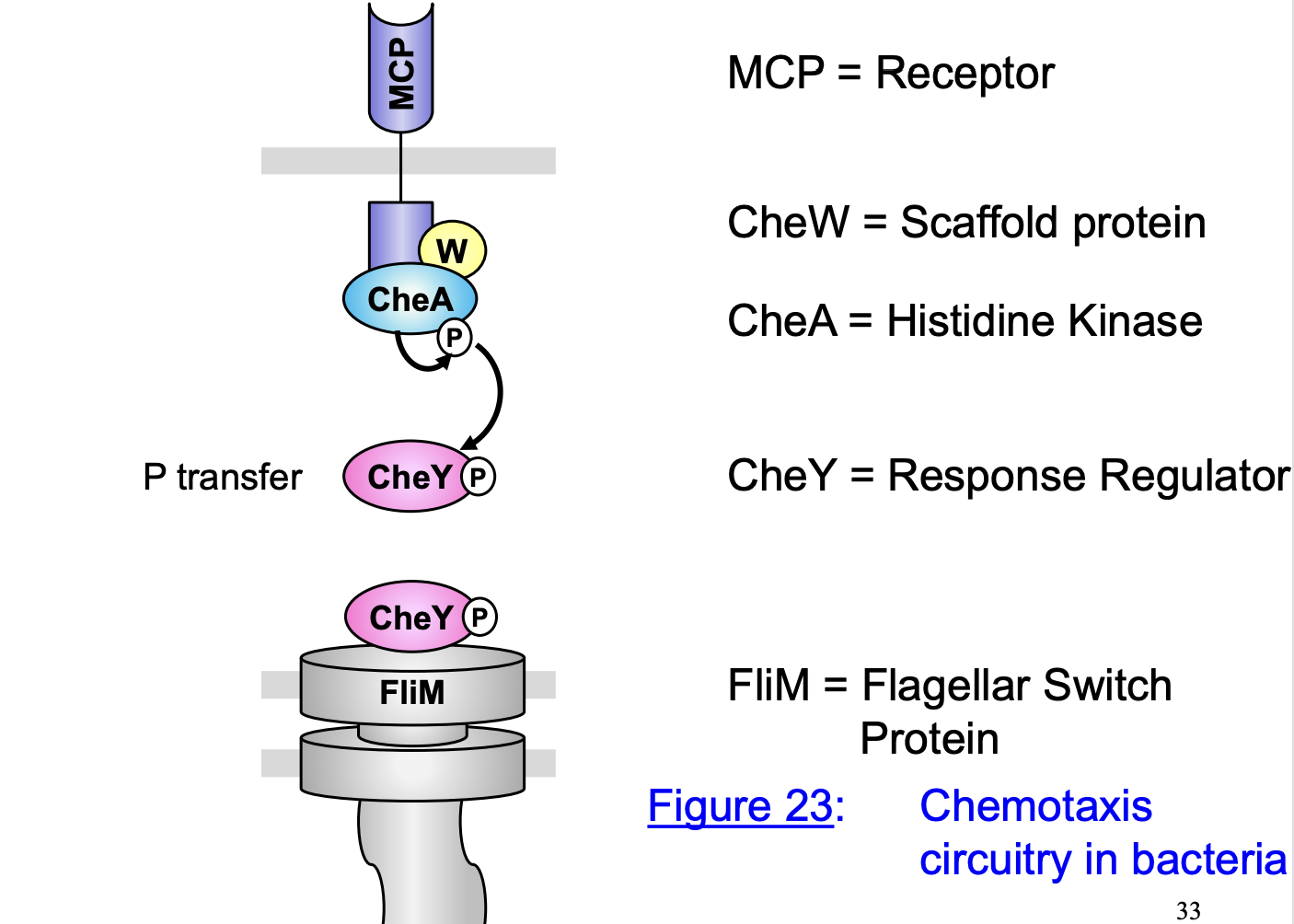
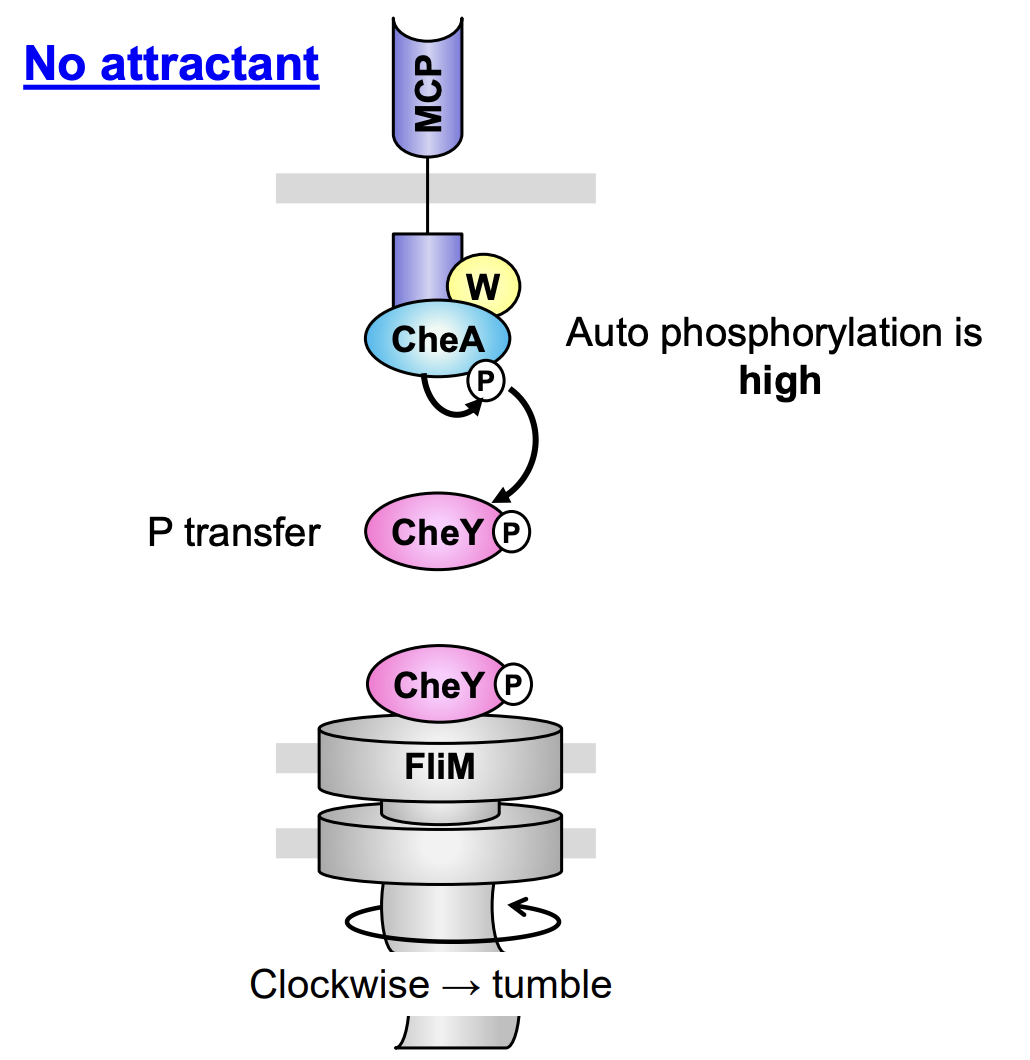
Chemotaxis transduction pathway A→ Attrctant absent/repellent present
CheW couple the receptor CheA
Due to lack of attractant, CheA = auto-phosphylated at His48
CheA-P transfers the P to CheY at Asp57 (transphosphylation of response regulator)
CheY-P interacts with the switch protein FliM at flagellar rotor
Flagella rotate clockwise and induce a tumble
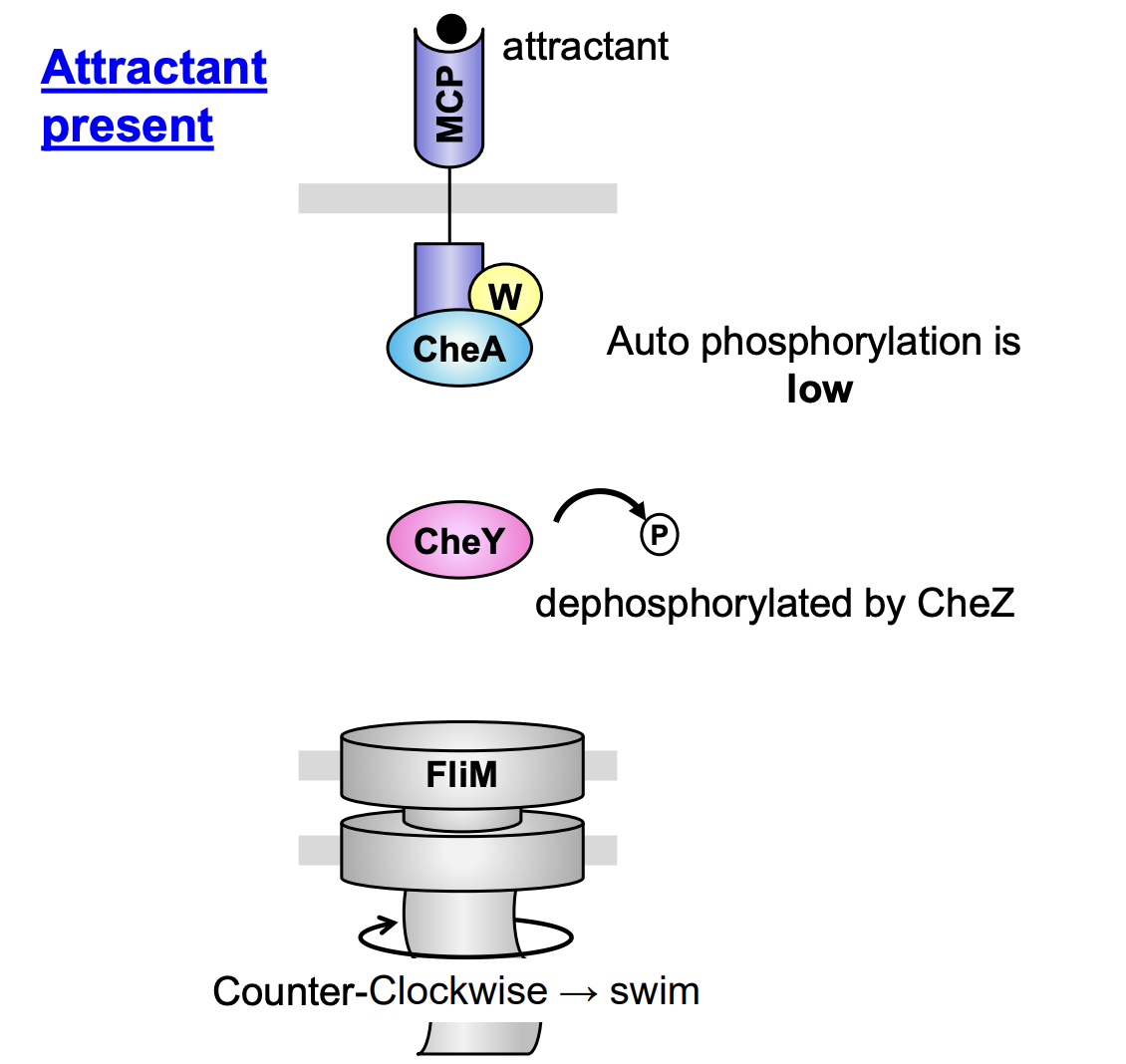
Chemotaxis transduction pathway B→ Attractant present
Attractants (AT) bind to chemotaxis receptor protein
Binding = low rate CheA autophosphylation
→ CheA is unphosphylated
Unphosphylated CheA cannot transfer the P to CheY
CheY also dephosphylated by CheZ
When CheY is dephosphylated, it cannot interact with FliM
→ flagella rotates counter-clockwise→ with long smooth runs
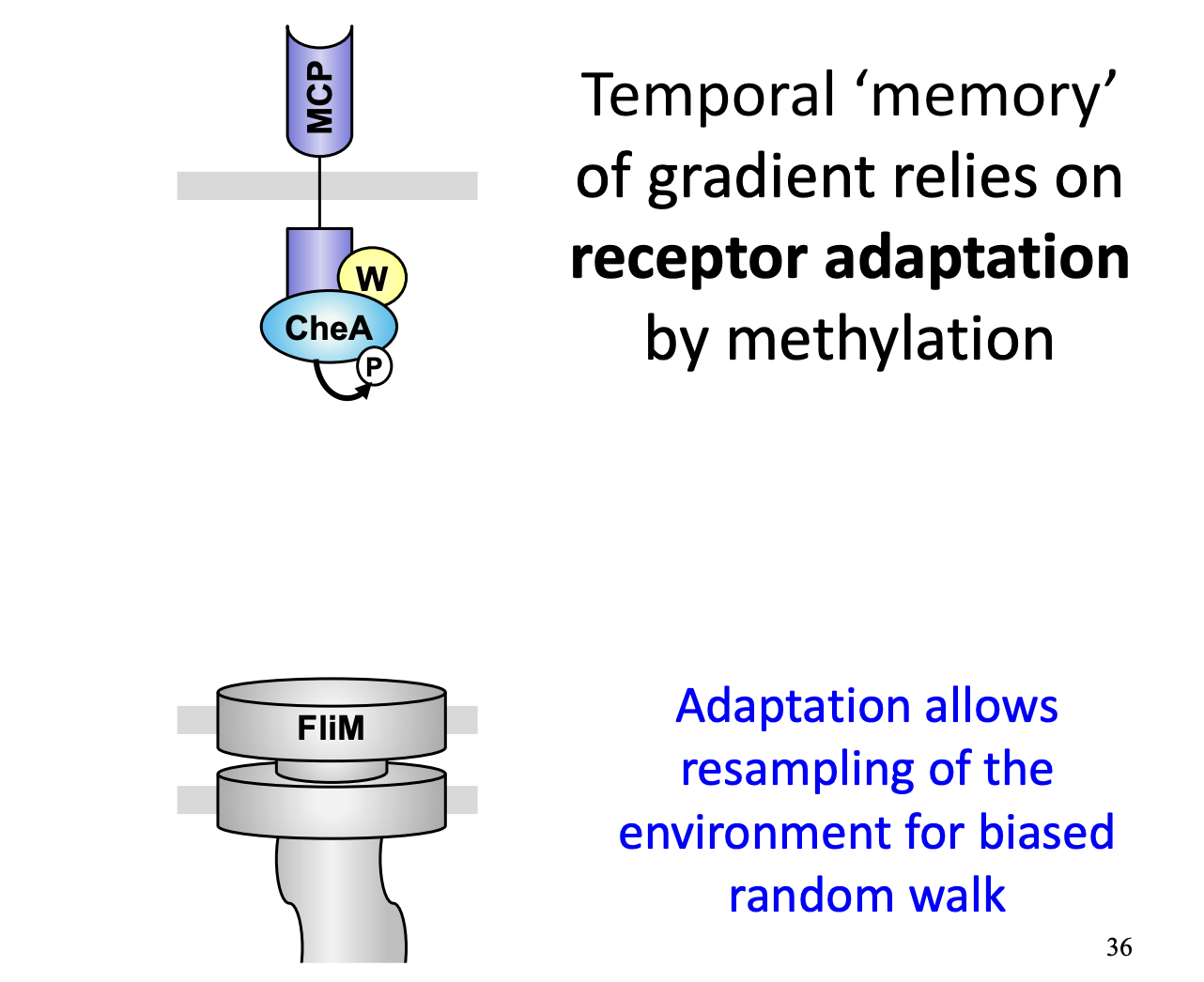
E.coli are too small to sense concentration differences at either end (coz like treacle). How do they detect concentration gradients?
use temporal memory
Achieved through a negative feedback mechanism→ based on methylation of MCP proteins
Uses the different timings of methylation and phosphorylation:
Methylation is slower than phosphorylation
Methylation is slower than de-methylation
Negative feedback mechanism→ based on methylation of MCP proteins
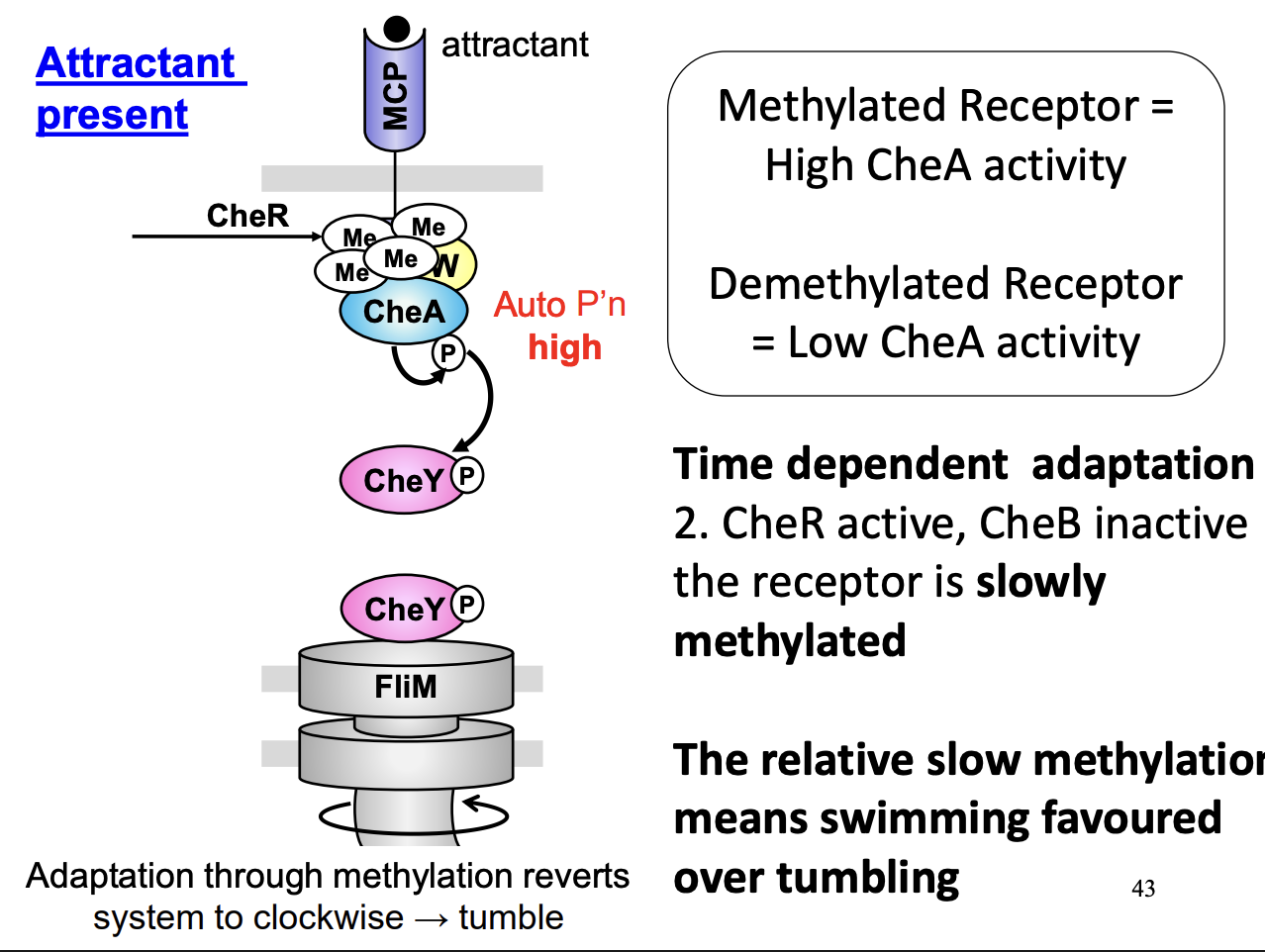
Chemotaxis transduction pathway C→ Adaptation
methylation of chemotaxis receptor protien
Acts as negative feedback→ returns system to an unstimulated state
CheR continuously methylates receptor whose response is influenced by degree of methylation state
CheB demethylating enzyme is determined by the phosphylation of CheA
Methylation of receptor CheR causes CheA stimulation and phosphylation
→ Methylation of receptor restores CheA activity→ following dephosphylation, caused by attractant binding to the receptor
Stimulation of CheA and phosphylation→ phosphylation and activity of CheB→ demethylation of the receptor
Causes→ restoration of CheA to dephosphylated low activity state
CheR→ constituively active
CheB→ regulated by CheA
These two types of protein modifications (de/phosphylation and de/methylation)
Occur at different timescales
→ methylation provides memory for system
→ essential for biased random walk
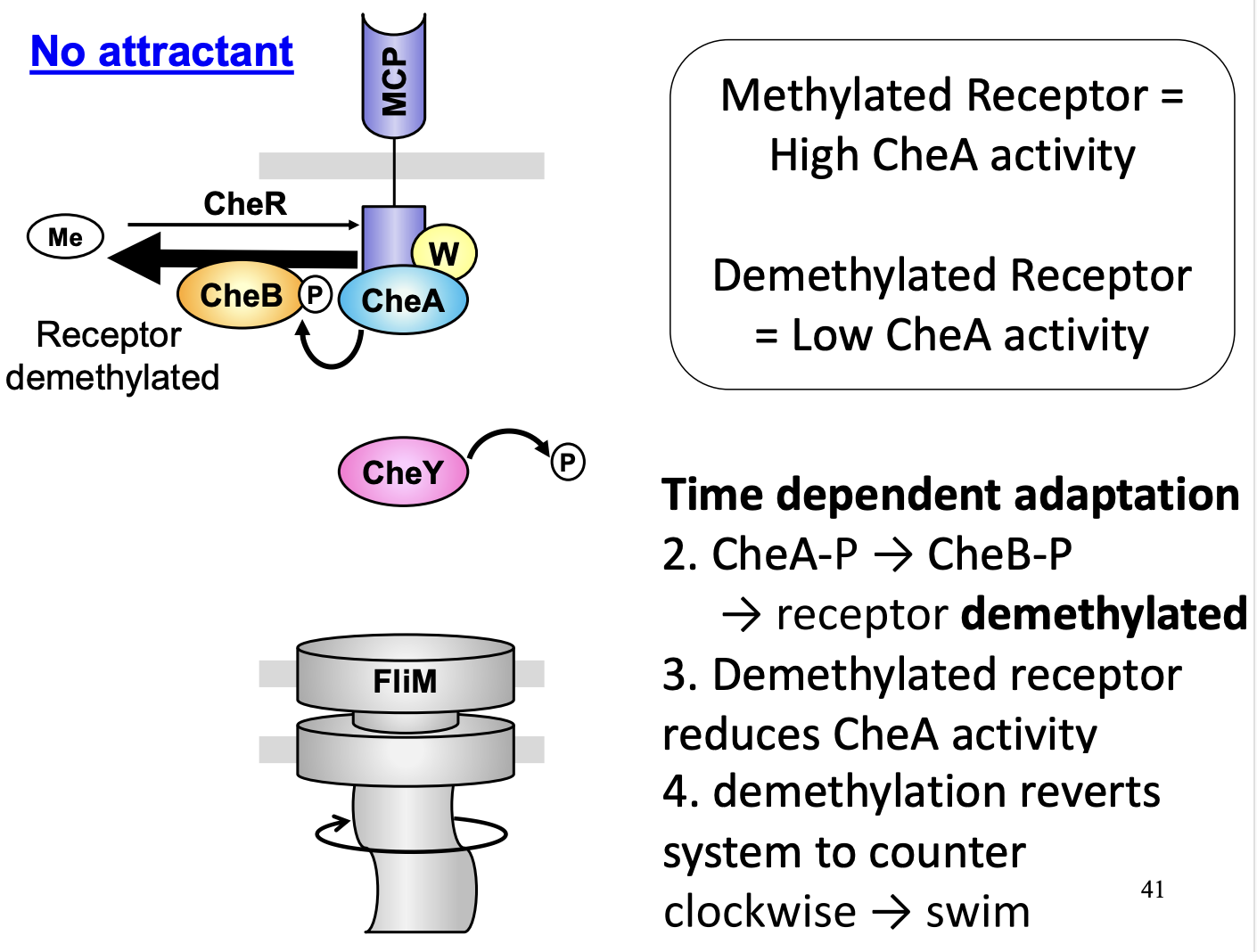
Temporal adaptation no attractant
NO attractant→ auto phophylation high→ CheA-P→ CheY-P= tumble
but
High CheA acitivty= CheB increased activity
CheB demthylated CheA
CheA now reduces activity
CheY dephosphylated by CheZ
→ swim
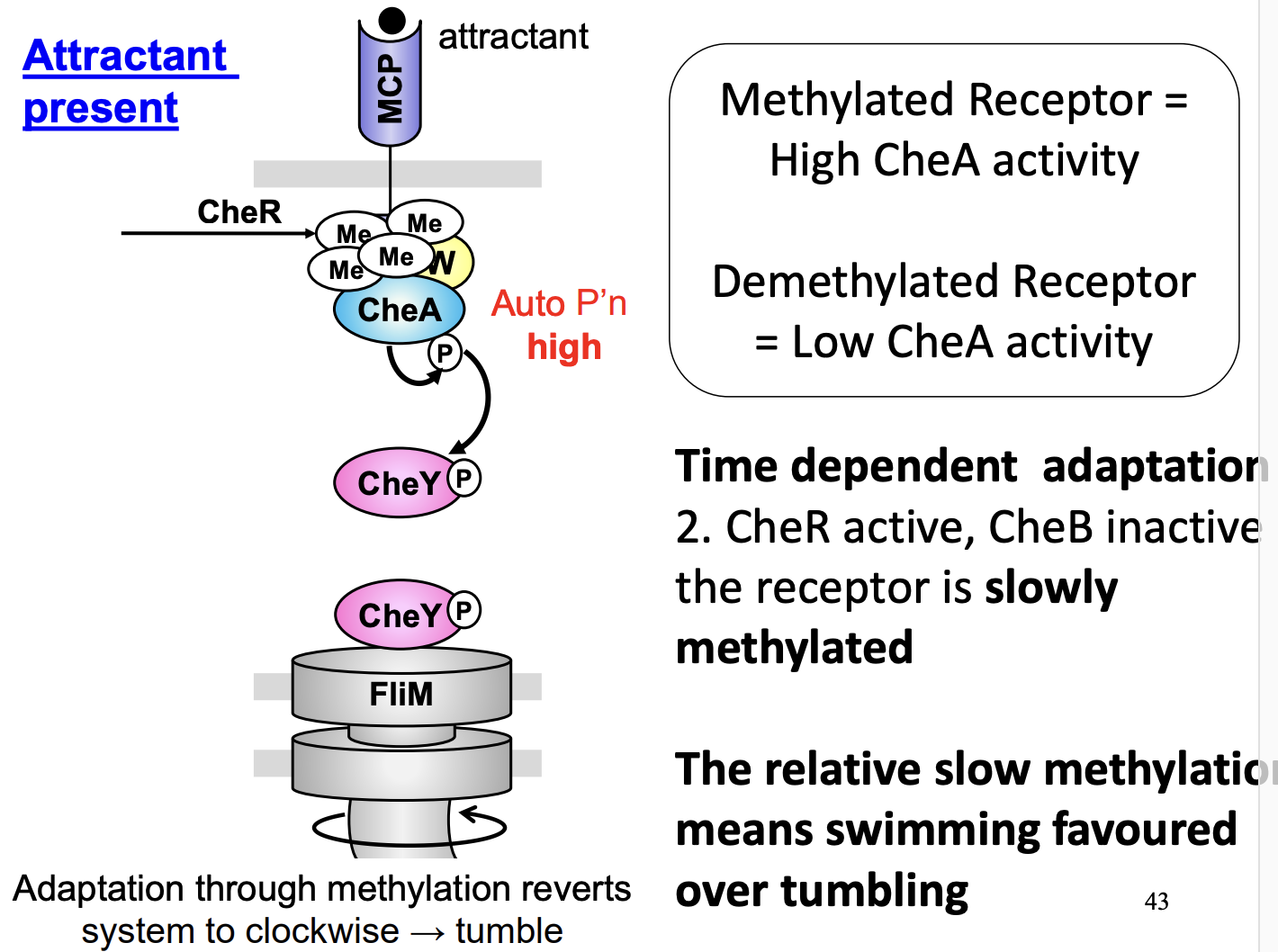
Temporal adaptation with attractant
Attractant= low autophophylation= Swimming
but
Consistent gradual methylation by CheR (and CheB inactive)
CheA methylated= high activity
Swimming favoured over tumbling
During attractant:
- because the methylation takes ages, so while it is waiting, the E.coli is swiming alot.
After methylation- > tumble
BUT
CheB de-methylation QUICKER→ so QUICKLY goes back to swimming
During no attractant→
same thing is happening→ promoting swimming
BUT→ constantly phosphylated→ so gets phospoylated back
goes BACK to tumbling
Self assessment questions
1. Why are there oscillations in the signal relay of aggregating slime molds?
2. What is quorum sensing?
3. What is a two component signaling system?
4. How do temporal delays contribute to chemotaxis in E. coli?