Eukaryotes Exam 4
1/181
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
182 Terms
self duplication (DNA replication)
H bonds between bases hold complementary strands together
each strand contains information required for construction of other strand
can act as template to direct synthesis of complementary strand and restore double-stranded state
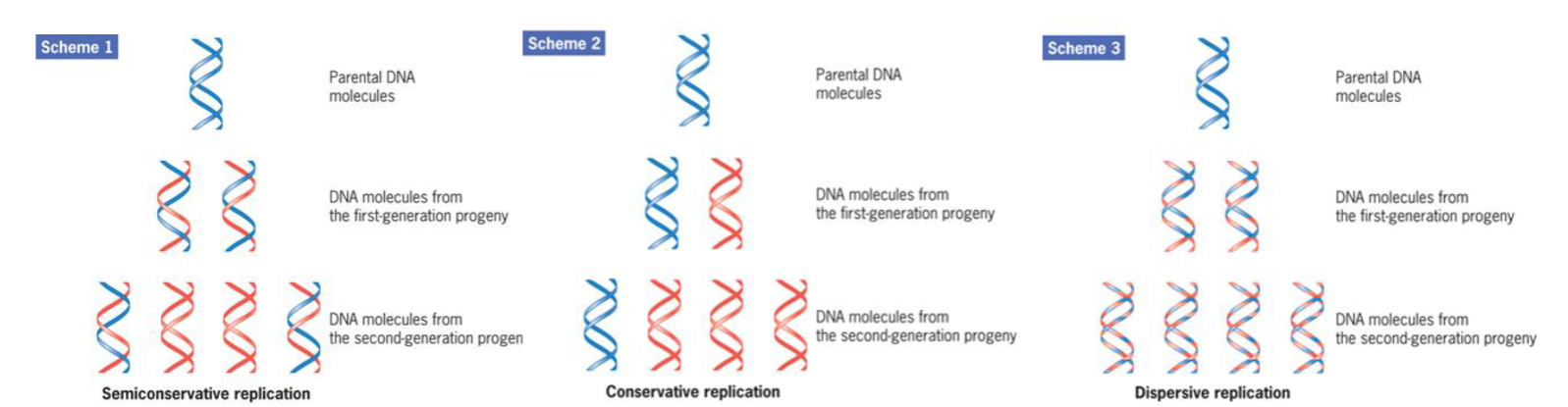
semiconservative
DNA replication is semiconservative because each daughter duplex contains one strand from the parent structure
Early progress in bacterial research was driven by 2 approaches:
isolation of temp‐sensitive (ts) mutants used to study DNA synthesis for replication, repair, and genetic recombination
In vitro studies with purified cellular components have shown activity of >30 proteins for E.coli chromosome replication
start of replication
starts at origin where proteins bind to initiate DNA replication, proceeds bidirectionally
Replication forks
sites where parental dsDNA helix unwinding
nucleotides being incorporated into newly synthesized strands
DNA Polymerases
responsible for synthesizing new DNA from DNA template
cannot initiate formation of DNA strand, requires primer
short strand provides 3′ OH terminus
adds nucleotides to 3′ hydroxyl terminus of existing strand
strand synthesis occurs 5'→3'
Semidiscontinuous Replication
both daughter strands synthesized simultaneously
DNA pol moves 3′ → 5′
2 newly assembled DNA strands grow in opposite directions, one growing toward replication fork, other growing away
major enzyme is DNA pol III
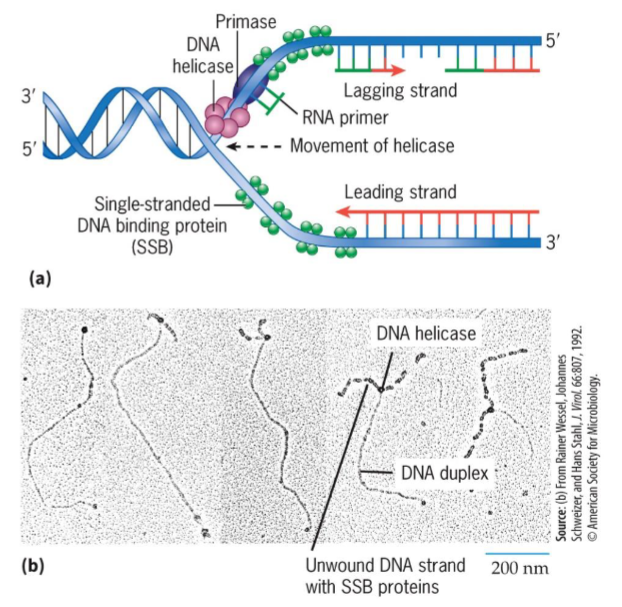
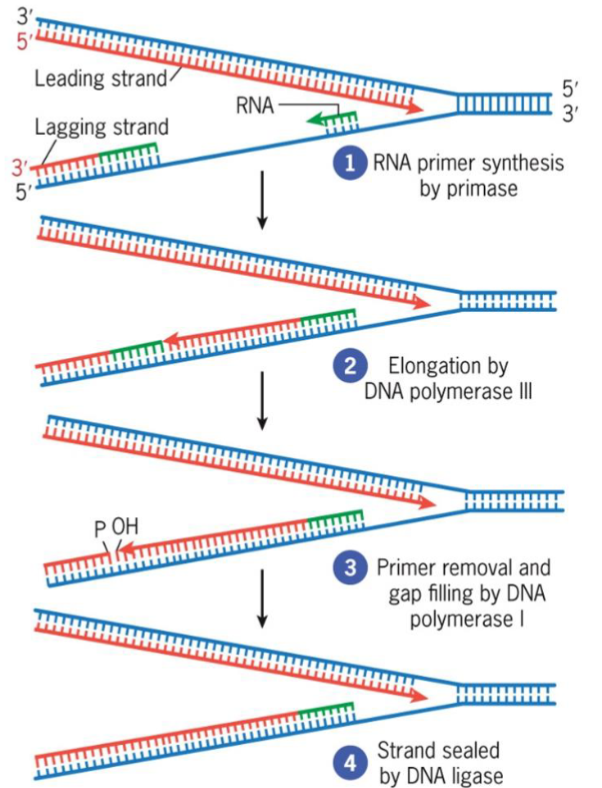
Primase
RNA polymerase that assembles short RNA primers
Okazaki fragments
small DNA segments
constructs lagging strand
uses DNA ligase to join into continuous strand
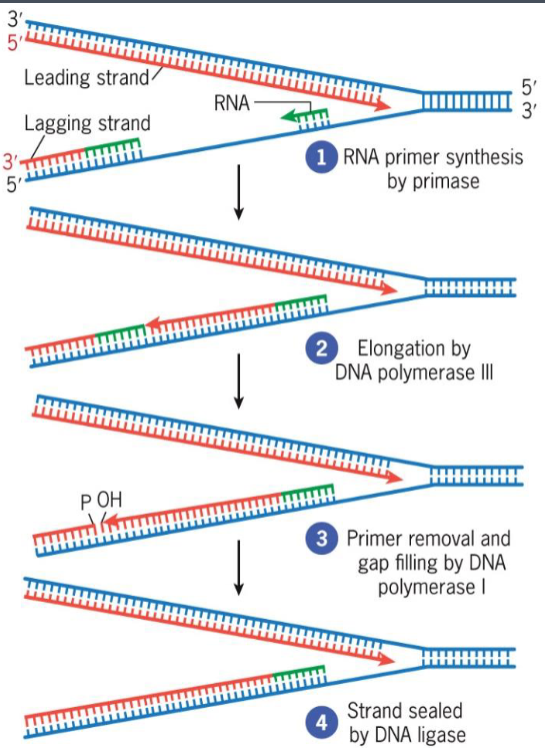
leading vs lagging strand synthesis
leading strand synthesized continuously, 5'→3'
lagging strand synthesized discontinuously, 5'→3 (constructed by Okazaki fragments)
Helicase
unwind a DNA duplex, uses energy from ATP hydrolysis
moves along one of DNA strands
breaks H bonds that hold 2 strands together
exposes single-stranded DNA templates
major helicase during replication in E. coli is DnaB
Single-stranded DNA-binding proteins (SSB)
stabilize single stranded DNA
DNA polymerase III holoenzyme
synthesize successive fragments of lagging strand
held to DNA by β clamp as it moves along template strand and synthesizes complementary strand
after completion of 1 Okazaki fragment, it disengages from β clamp
cycles to recently assembled clamp “waiting” at upstream RNA primer–DNA template junction, forms another Okazaki fragment
original β clamp left behind for period on finished Okazaki fragment, eventually disassembled and reutilized
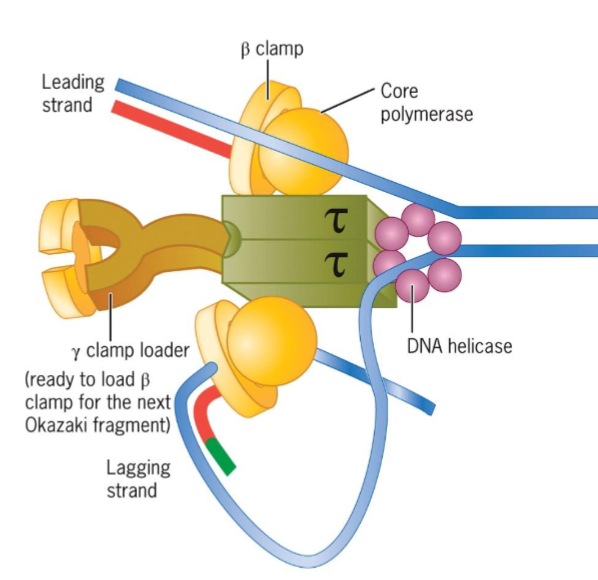
β clamp
keeps polymerase associated with DNA
DNA polymerase I
involved in DNA repair
removes RNA primers at 5′ end of Okazaki fragments during replication and replaces with DNA
maximizes fidelity of DNA replication
5′ → 3′ and 3′ → 5′ exonucleases
polymerase activity simultaneously fills gap with deoxyribonucleotides
uses DNA ligase to seal gaps between fragments
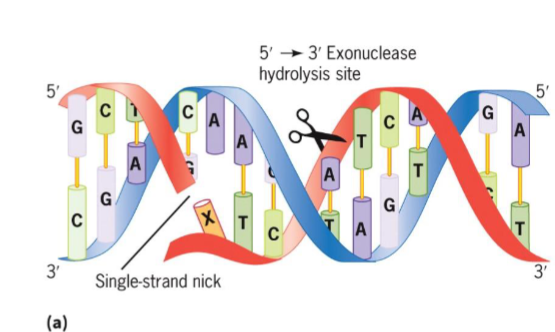
5′ → 3′ exonuclease (DNA pol I)
removes nucleotides from 5′ end of single-strand nick
RNA primers removal
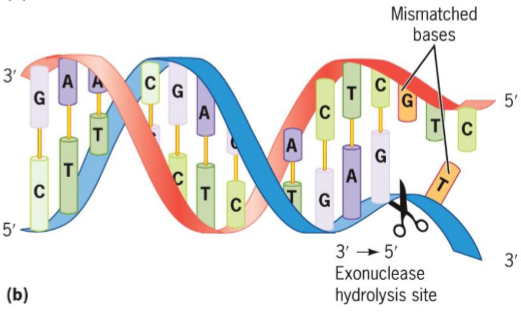
3′ → 5′ exonuclease
removes mispaired nucleotides from 3′ end of growing DNA strand
DNA repair
3 experimental systems that help understanding of replication in euks:
isolation of mutant yeast and animal cells unable to produce specific gene products required for various aspects of replication
analysis of structure and mechanism of action of homologous replication proteins from archaeal species
development of in vitro systems using cell extracts or purified proteins
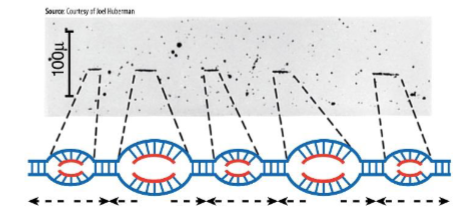
replicons
small portions of eukaryotes genomes for replication
has its own origin (and ARS) from which replication forks proceed outward in both directions
Autonomous replicating sequence (ARS)
associated with multiprotein complex called origin recognition complex (ORC)
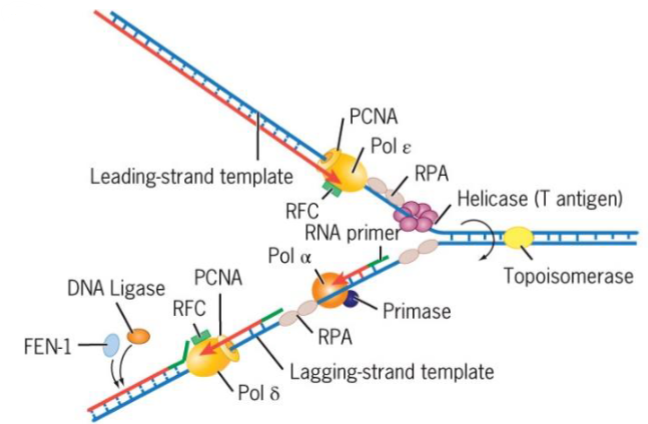
5 polymerases of Eukaryotic Replication Fork
α - initiates Okazaki fragment synthesis
β - involved in DNA repair
γ replicates mtDNA
δ (delta) - lagging strand synthesis
ε - leading strand synthesis
DNA is synthesized semidiscontinuously
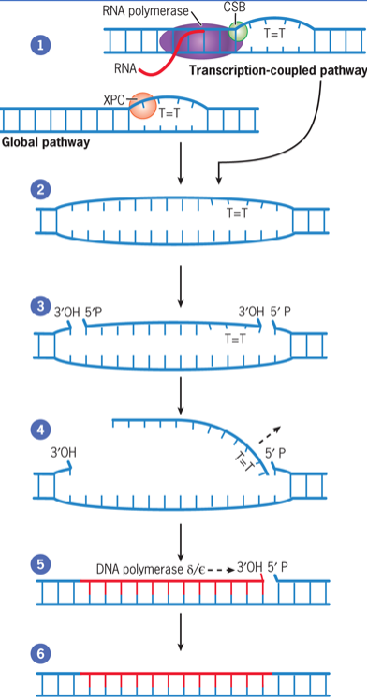
Nucleotide Excision Repair (NER)
cut- and-patch mechanism that removes bulky lesions
generated by environmental mutagens such as UV irradiation and bulky chemical compounds
two distinct pathways: transcription-coupled and global genomic paths
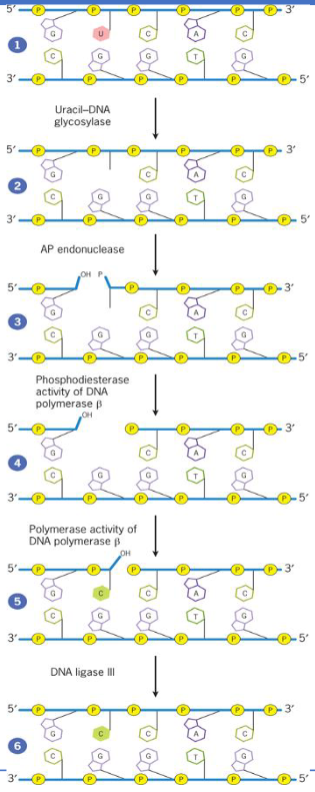
Base Excision Repair (BER)
removes altered nucleotides generated by reactive chemicals from diet or metabolism
corrects only the damaged bases
initiated by DNA glycosylase which recognizes and removes altered bases by cleavage of glycosidic bond holding base to deoxyribose sugar
Mismatch repair (MMR)
correction of mistakes that escape DNA polymerase proofreading activity
repair enzymes recognize distortions caused by mismatched bases
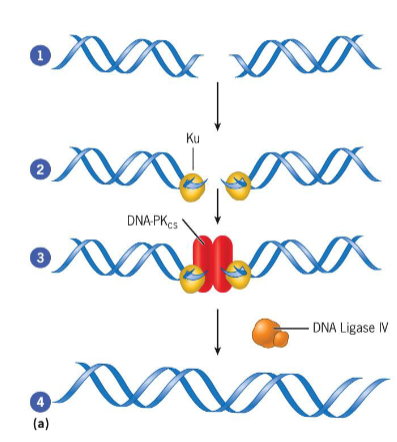
Double Strand Break (DSB) repair
nonhomologous end joining (NHEJ): complex of proteins binds to broken ends of DNA duplex and catalyzes reactions that rejoin broken strands
homologous recombination: requires homologous chromosome to serve as template for repair of broken strand
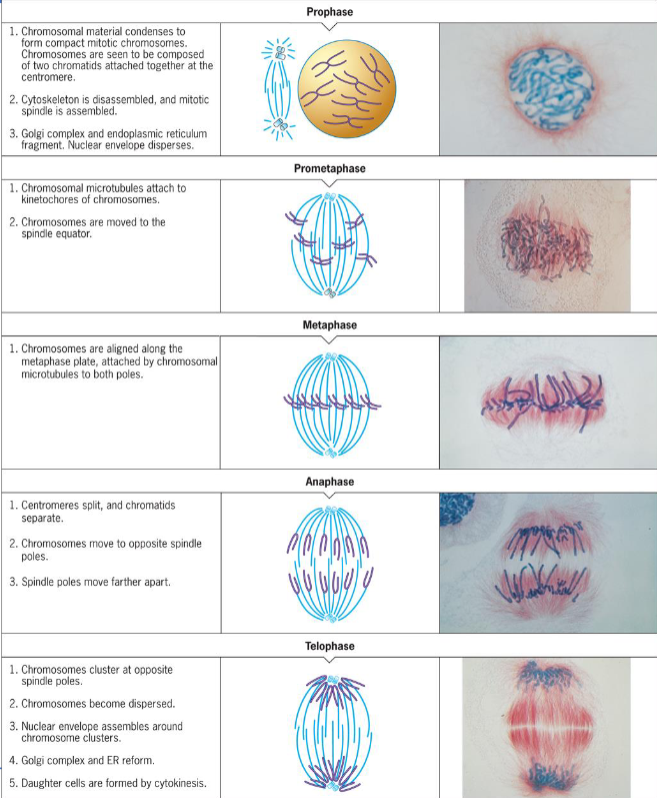
Mitosis
production of cells that are genetically identical to parent
basis for producing new cells
process of nuclear division in which two nuclei are produced to maintain chromosome number
consists of Prophase, Prometaphase, Metaphase, Anaphase, Telophase which represents a segment of a continuous process
Meiosis (reduction)
production of cells with half genetic content of parent
basis for producing new sexually reproducing organisms
ensures production of haploid phase in life cycle, and fertilization ensures diploid phase
without this, chromosome numbers would double each generation, making sexual reproduction unsustainable
gametic, zygotic, sporic
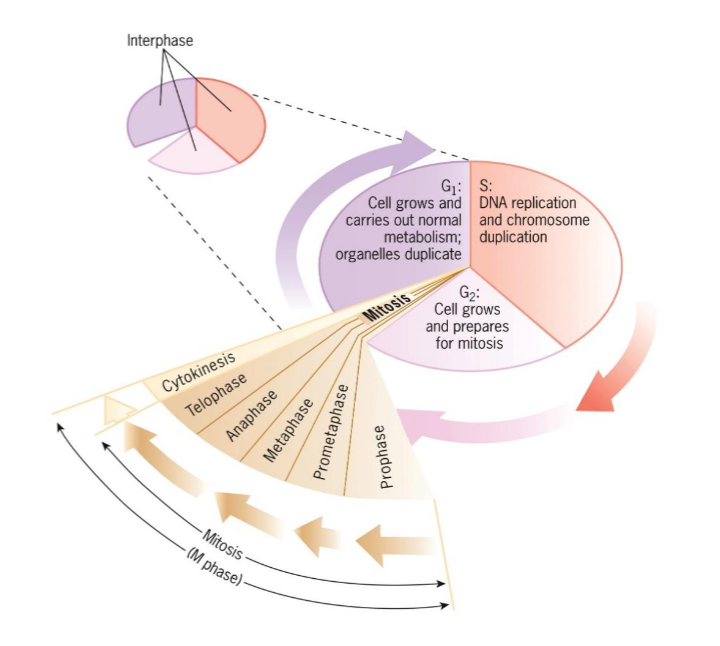
Cell Cycle Stages
Interphase
G1
S
G2
M phase
Mitosis
Prophase
Metaphase
Anaphase
Telophase
Cytokinesis
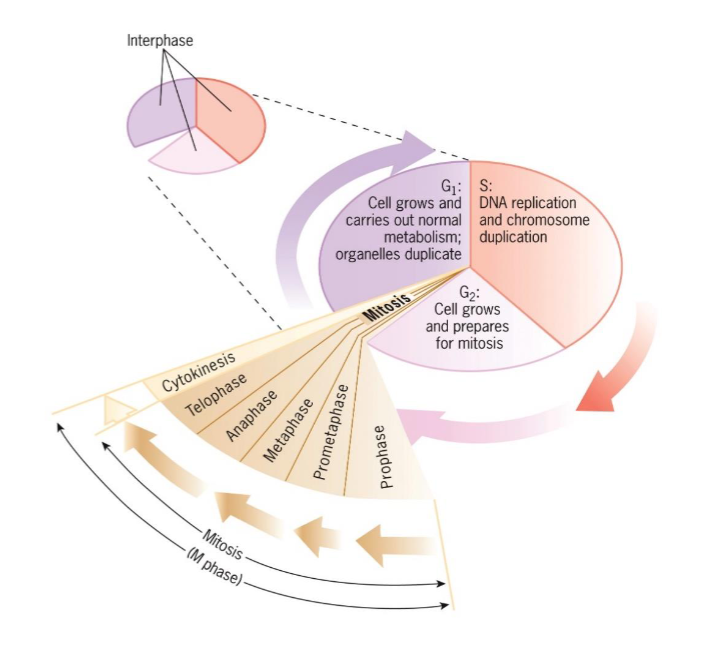
M phase
consists of mitosis and cytokinesis
Cell contents divide into two new cells
Mitosis lasts about an hour
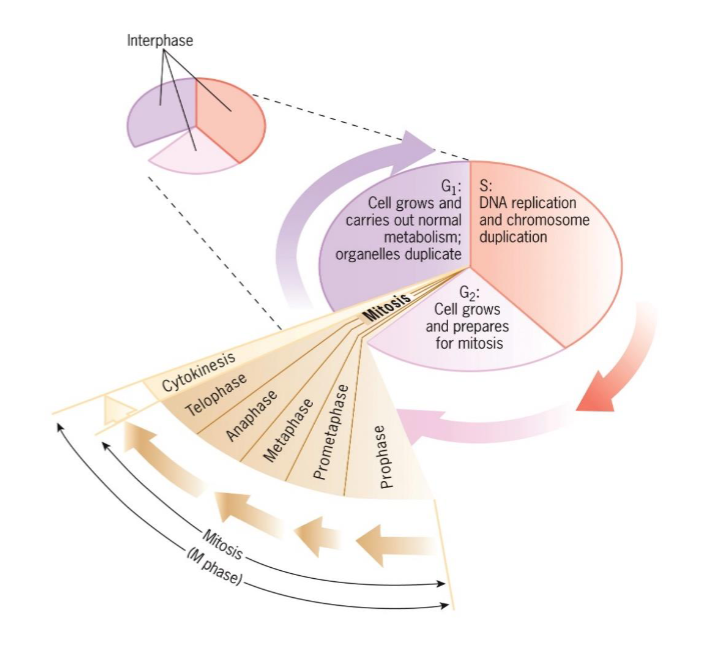
Interphase
G1, S, G2
period between cell divisions
cell grows and engages in diverse metabolic activities
lasts longer than M phase; (day-week+)
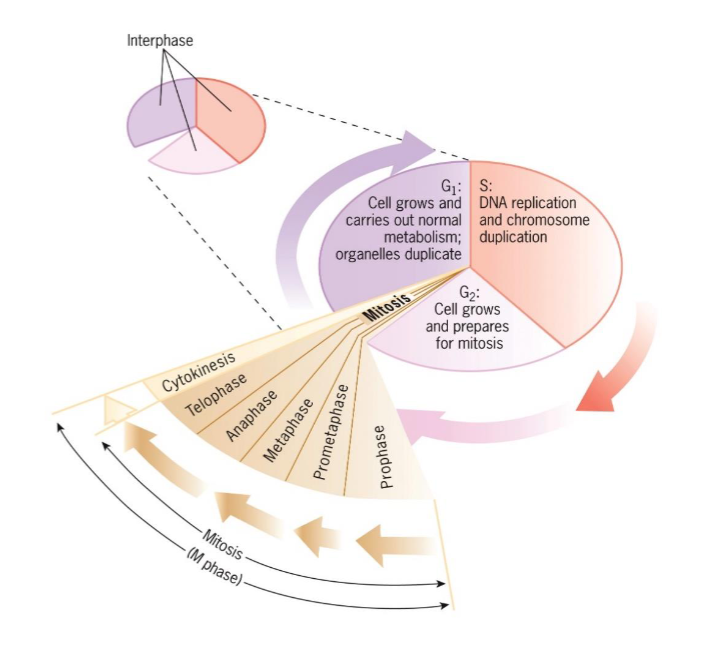
G1 (interphase)
takes place between end of mitosis
cell growth and carries out normal metabolism and organelles duplication
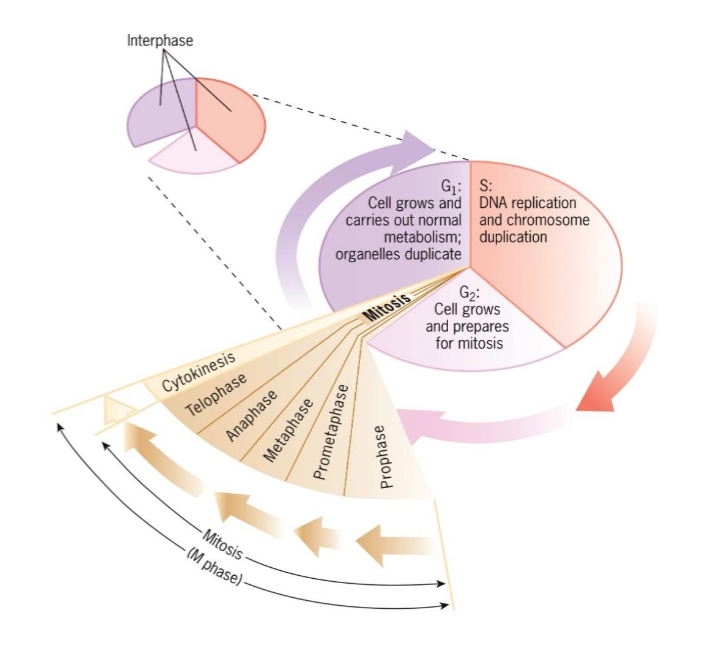
S phase (interphase)
DNA replication and chromosome duplication
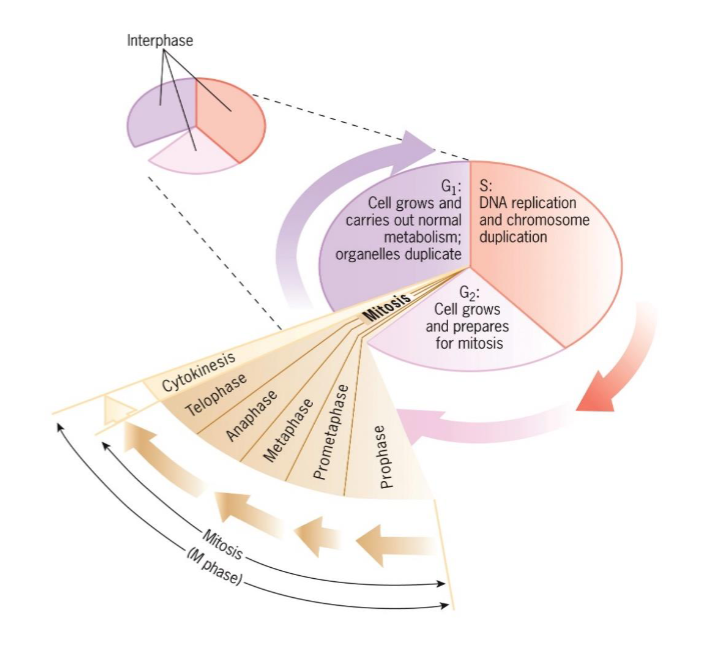
G2 (interphase)
occurs between end of S and beginning of mitosis
cell grows and prepares for mitosis
3 cell types with capacity to grow and divide in vivo
Cells that are highly specialized and lack ability to divide. (nerve cells, muscle cells, or red blood cells)
Cells that normally don’t divide but can be induced to begin DNA synthesis and divide when given stimulus. (liver cells and lymphocytes)
Cells that normally possess relatively high level of mitotic activity. (stem cells for blood elements, skin and other epithelia, plant apical meristems)
maturation-promoting factor (MPF)
activation triggers entry into M phase
two subunits: kinase and cyclin (regulatory subunit)
increased concentration of cyclin activates kinase
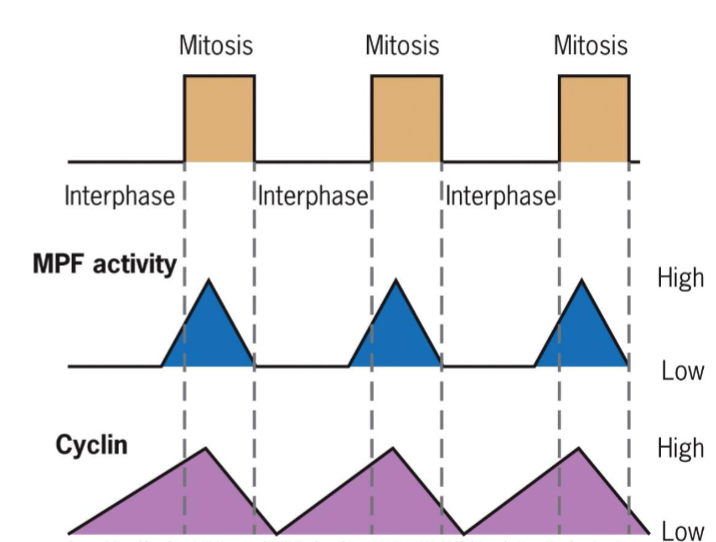
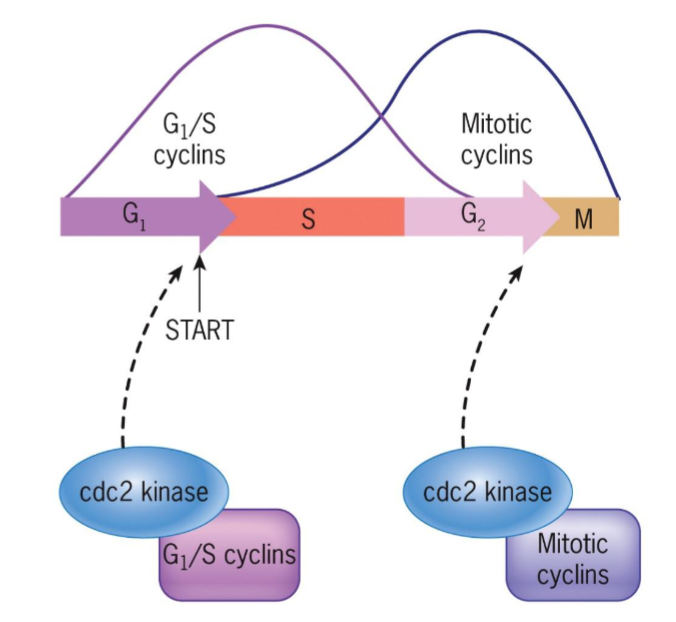
cyclin-dependent kinases (Cdks)
MPF-like, studied in yeast Ts mutants
products of cdc2 (fission yeast) and cdc28 (budding yeast) genes responsible for passage through G1 (START) and G2 control points
START – cell committed to DNA replication (G1/ S cyclins)
G2/M transition (Mitotic cyclins)
Checkpoints
check if chromosomal DNA is damaged
check if certain critical processes such as DNA replication during S phase or chromosome alignment during M phase, have not been properly completed
progress through cell cycle can be arrested at checkpoint by sensors that detect chromosomal abnormalities, transmitters that signal the information, effectors that inhibit cell cycle machinery
prophase (mitosis)
formation of mitotic chromosome which consists of two chromatids
mitotic machinery assembled
duplicated chromosomes prepared for segregation
chromosome compaction/condensation occurs in early prophase
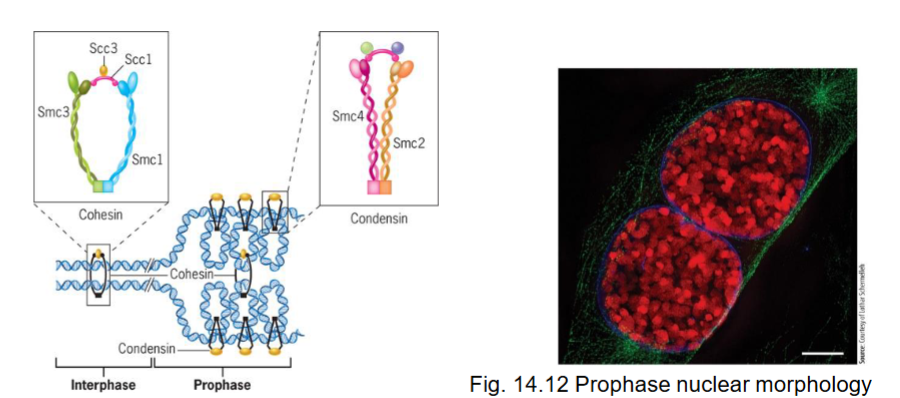
Cohesin
with condensin, are proteins responsible for compaction
associates with DNA of each chromosome prior to replication, forms ring to encircle 2 sister DNA molecules
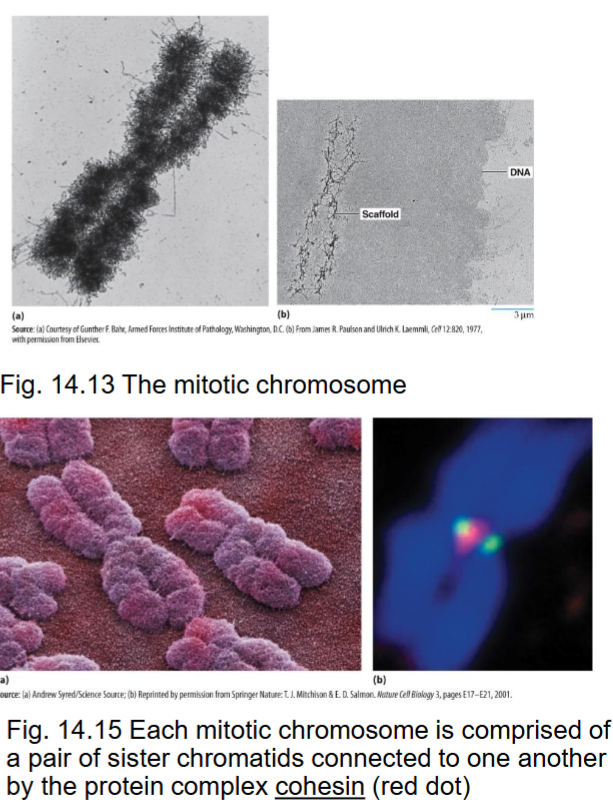
Centromeres
occur at a primary constriction on chromosomes and serve as binding site for proteins
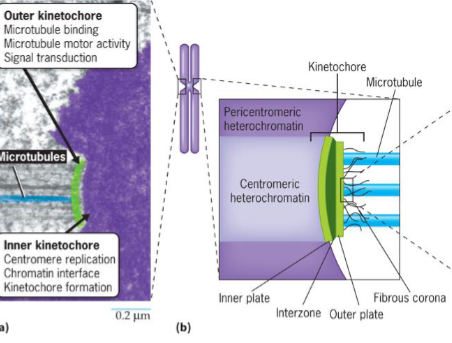
Kinetochores
on outer surface of centromeres
sites where chromosomes attach to microtubules of mitotic spindle
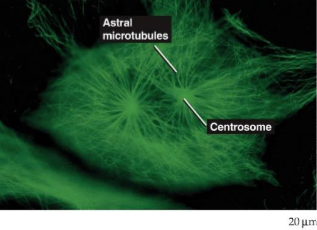
mitotic spindle
entering mitosis, microtubules undergo disassembly before reassembly into microtubules
in animal cells, microtubules are arranged in aster around each centrosome
Nuclear pore complexes
disassembled, nucleoporin subcomplexes disrupted and subcomplexes dissociate
Nuclear lamina
is disassembled - depolymerization of lamin filaments
Nuclear membranes
are disrupted mechanically - holes are torn into nuclear envelope by cytoplasmic dynein molecules
Membranous organelles (mitochondria, lysosomes, and peroxisomes)
remain relatively intact through mitosis
Golgi complex
may become incorporated into ER during prophase or become fragmented to form distinct population of small vesicles partitioned between daughter cells
Prometaphase (mitosis)
mitotic spindle assembly completed
chromosomes moved by microtubules into center of cell
single kinetochore attached to microtubules form both spindle poles
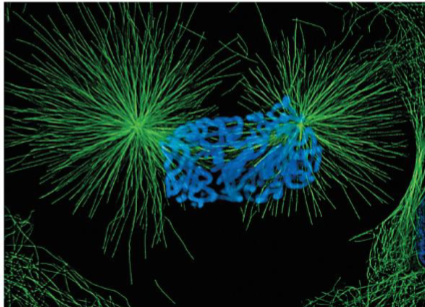
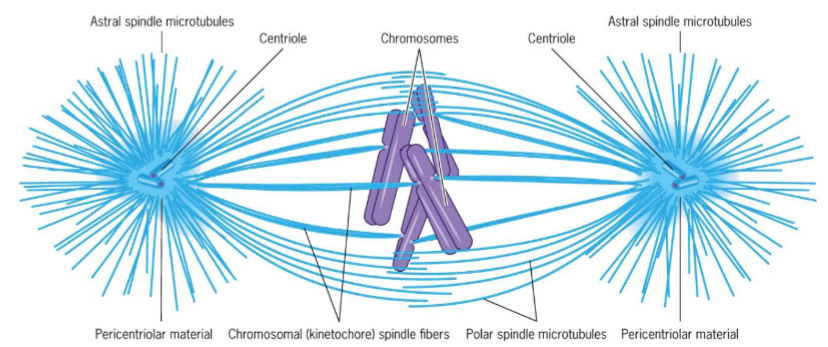
Metaphase (mitosis) + 3 microtubule groups
Astral microtubules - radiate from centrosomes to regions outside spindle body, help position spindle apparatus and determine plane of cytokinesis
Chromosomal spindle fibers - exert pulling force on kinetochores maintaining chromosome in equatorial plane
Polar microtubules - maintain integrity of spindle
chromosomes aligned at spindle equator on metaphase plate
Anaphase (mitosis)
begins when sister chromatids of each chromosome split apart and start their movement toward opposite poles
Anaphase A and B: chromosomes split in synchrony
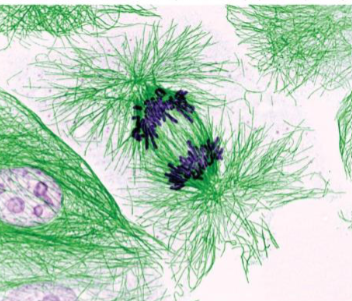
Anaphase A (mitosis)
movement of chromosomes toward poles
tubulin subunits lost from both ends of chromosomal microtubules, resulting in shortening and movement of chromosomal fibers
Anaphase B (mitosis)
2 spindle poles move in opposite directions due to elongation of microtubules
tubulin subunits added to + ends of polar microtubules
Telophase (mitosis)
final stage of mitosis, daughter cells return to interphase
mitotic spindle disassembles
nuclear envelopes of 2 nuclei reassembled
chromosomes become dispersed
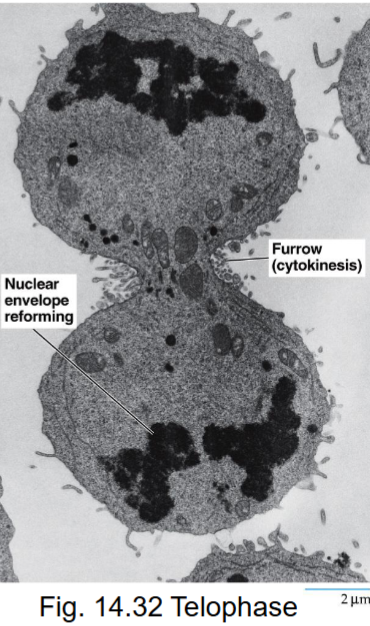
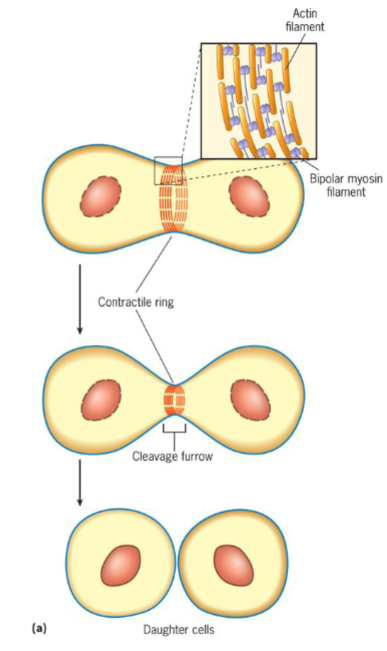
Cytokinesis (M phase)
process where 1 cell is divided into 2 daughter cells
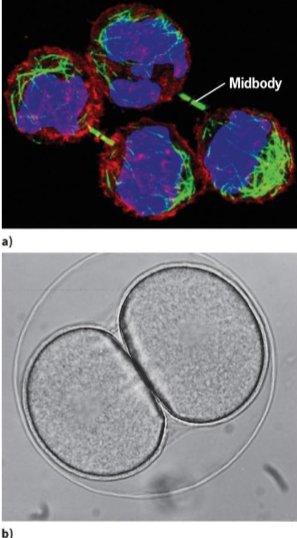
contractile ring theory
suggests that a thin band of actin and myosin filaments generates force to cleave cell
site of filament assembly (plane of cytokinesis) determined by signal coming from spindle poles
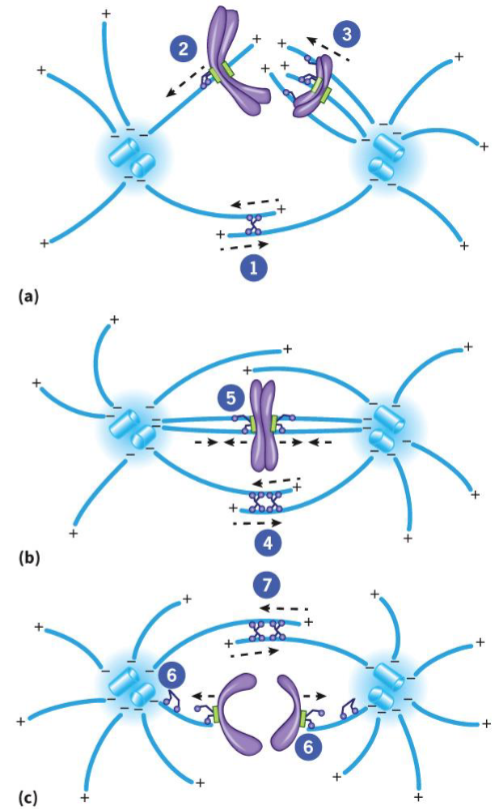
Motor Proteins
powered by microtubule motors (dynein and kinesin-related proteins)
located at spindle poles and kinetochores, keeps poles apart
bring chromosomes to metaphase plate and keep them there
elongate the spindle during anaphase B
2 unique challenges plant cells face when undergoing division
rigid cell wall and little freedom to move
Preprophase band
dense ring of cytoskeleton filaments and proteins
microtubules, actin, organelles, accessory proteins
band disappears by prometaphase, but cell remembers its original location
division plane forms at former site
molecular mark is unknown, membrane proteins recruited and retained at site during mitosis
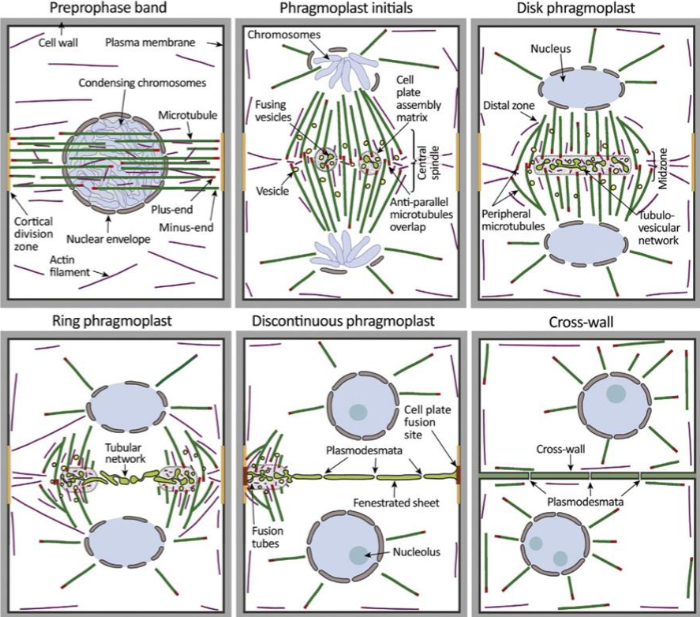
cell plate
precursor structure that allows plant cells to build new extracellular wall during division
starts from cell center and growing outward
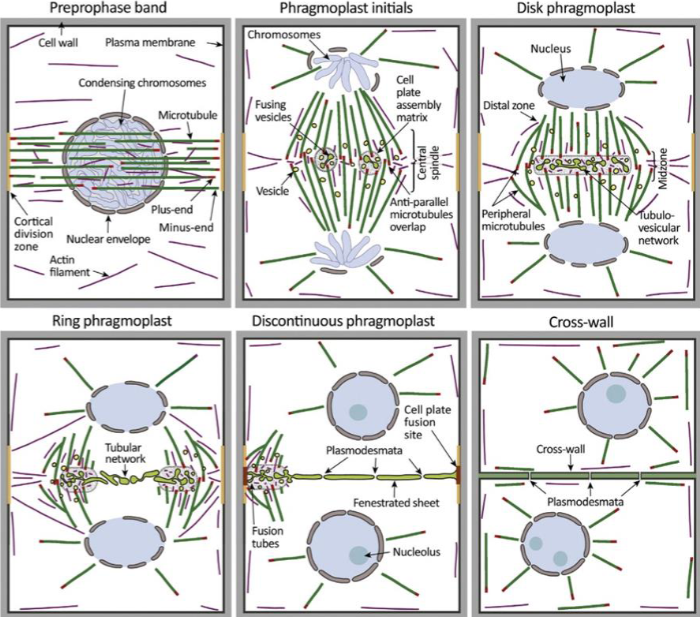
phragmoplast
guides cell plate formation
during cytokinesis, remnants of central spindle transport Golgi-derived vesicles to midzone
made of vesicles, cytoskeletal proteins and membranes
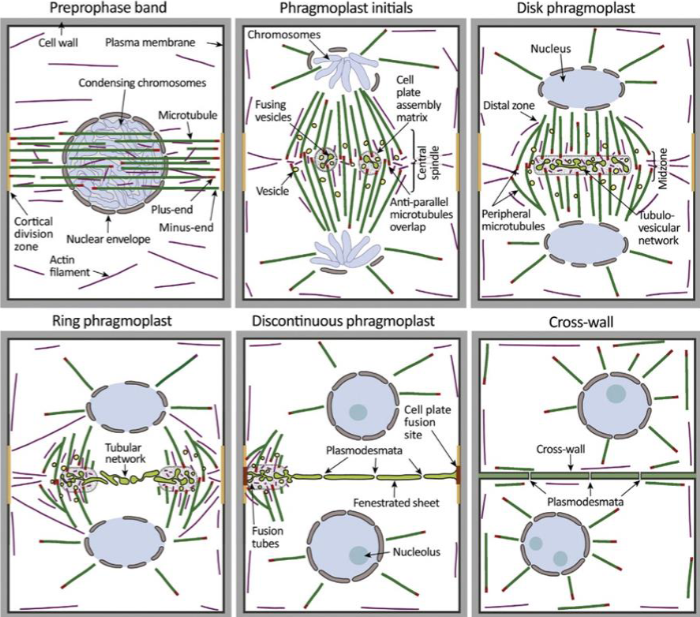
plant cell wall formation
vesicles fuse midzone to form tubular, disk-shaped membrane network
as phragmoplast expands to cell edges, cell wall matures and fuses with parent plasma membrane, forming cell wall
requires gradual deposition of polysaccharides, pectin, cellulose, and hemicellulose into cell wall lumen
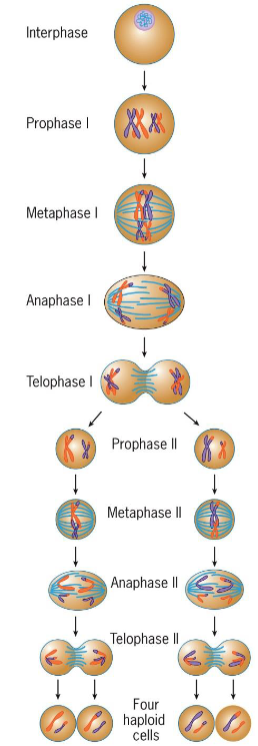
meiosis stages
interphase
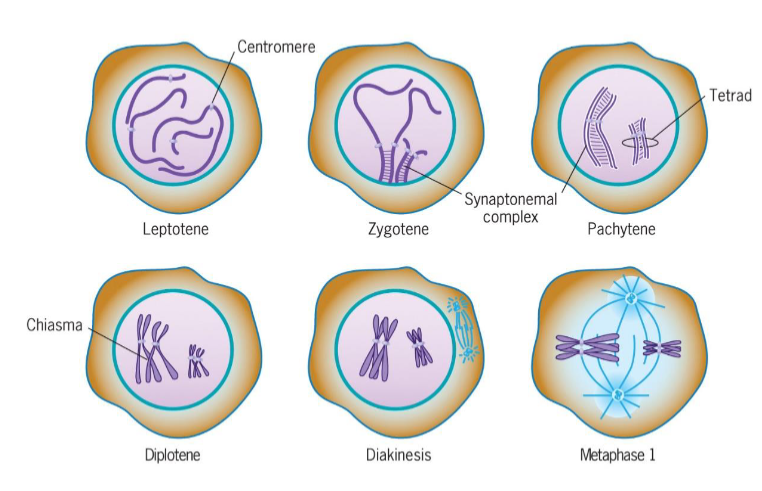
Prophase 1
leptotene
zygotene
pachytene
diplotene
diakinesis
MAT 1
PMAT 2
4 haploid cells
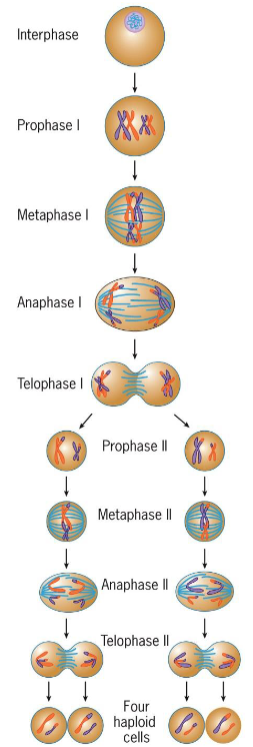
Meiosis I
homologous chromosomes pair and then segregate, ensuring that daughter cells receive full haploid set of chromosomes
Genetic recombination takes place
Start with diploid parent cells and end with 2 haploid daughter cells
Meiosis II
sister chromatids separated, simpler then meiosis 1
start with 2 haploid parent cells and end with 4 haploid daughter cells, maintaining # of chromosomes and DNA in each cell
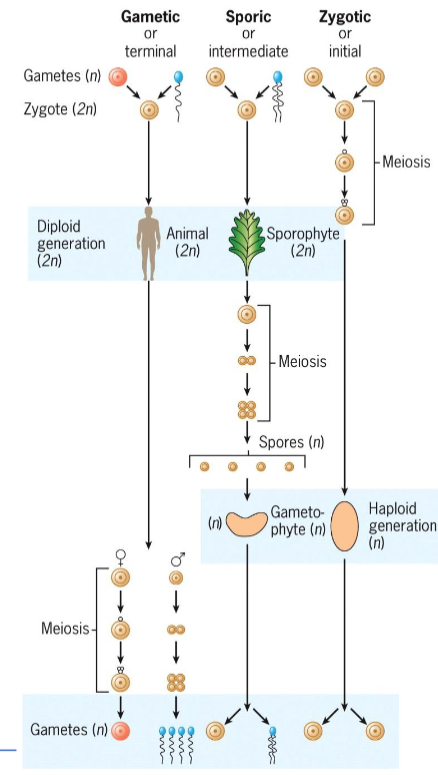
Gametic (terminal) meiosis
in animals and protists
meiosis occurs during gamete formation
in males, spermatogonia become spermatocytes → meiosis → spermatids → sperm
in females, oocytes enter extended meiotic prophase; meiosis completes only after fertilization
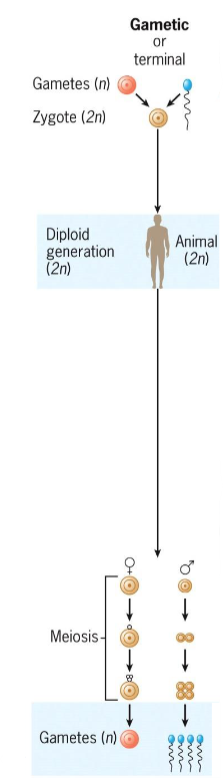
Zygotic (initial) meiosis
found in protists and fungi
meiosis happens immediately after fertilization, forming haploid spores
spores undergo mitosis to produce haploid adults; diploid phase is brief

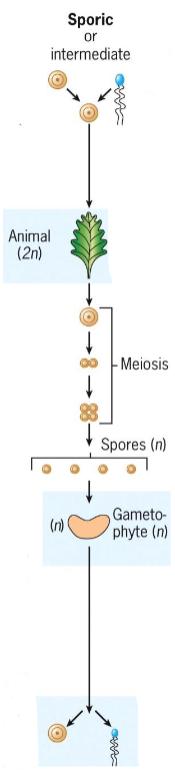
Sporic (intermediate) meiosis
Occurs in plants and algae
Meiosis occurs during sporogenesis in diploid sporophyte
Spores grow into haploid gametophytes, which produce gametes via mitosis
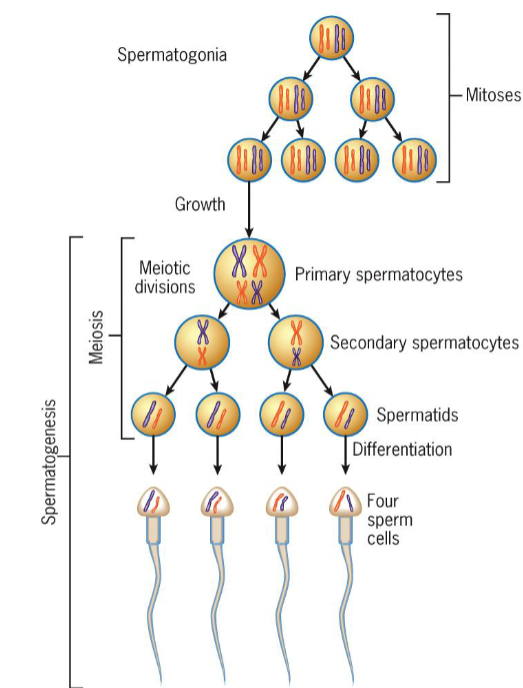
Gametic Meiosis - Male
meiosis occurs prior to differentiation of spermatozoa
2 divisions of meiosis produces 4 undifferentiated spermatids
each spermatid undergoes complex differentiation to become highly specialized sperm cell
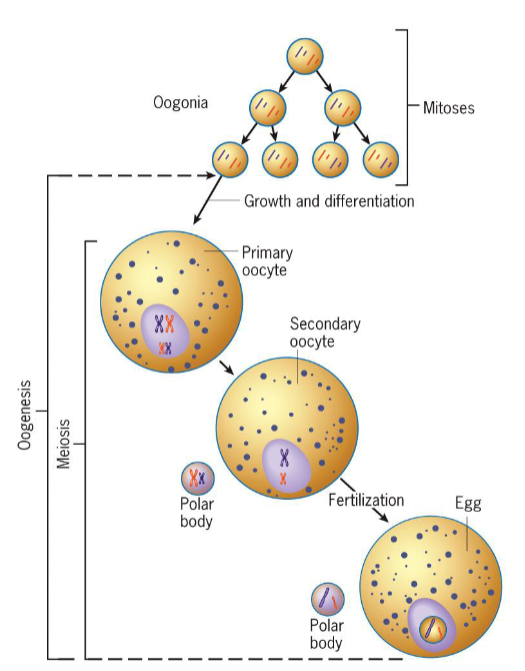
Gametic Meiosis - Female
oogonia become primary oocytes, which then enter extended meiotic prophase
vertebrate eggs fertilized before completion of meiosis (usually metaphase II)
meiosis completed after fertilization, while sperm resides in egg cytoplasm
only after differentiation of oocyte is complete does meiotic divisions occur
Prophase 1 (meiosis)
DNA replicated prior to meiosis
consists of leptotene, zygotene, pachytene, diplotene, diakinesis
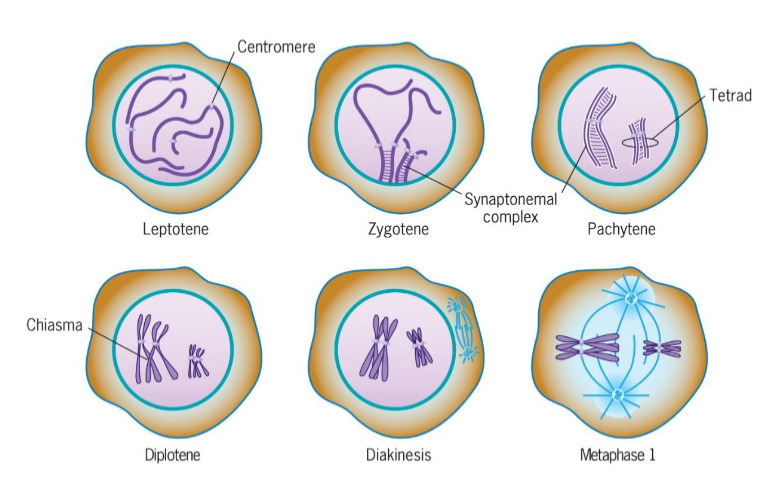
Leptotene (Prophase 1 of meiosis)
chromosomal condensation starts
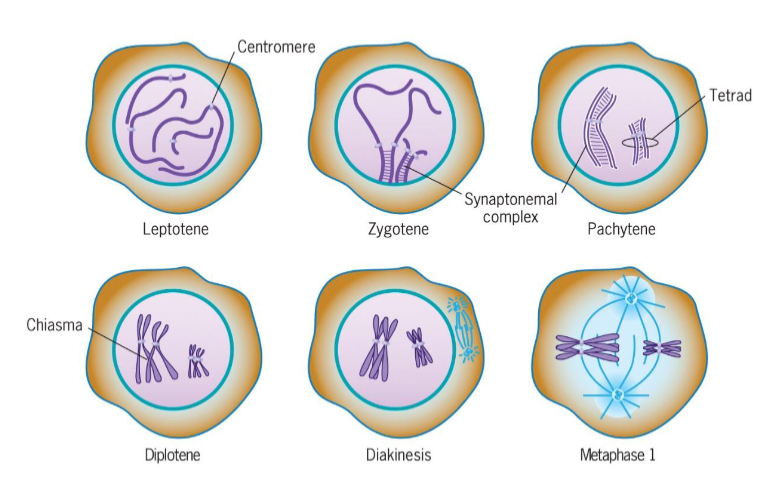
Zygotene (Prophase 1 of meiosis)
synapsis, homologous chromosomes pair
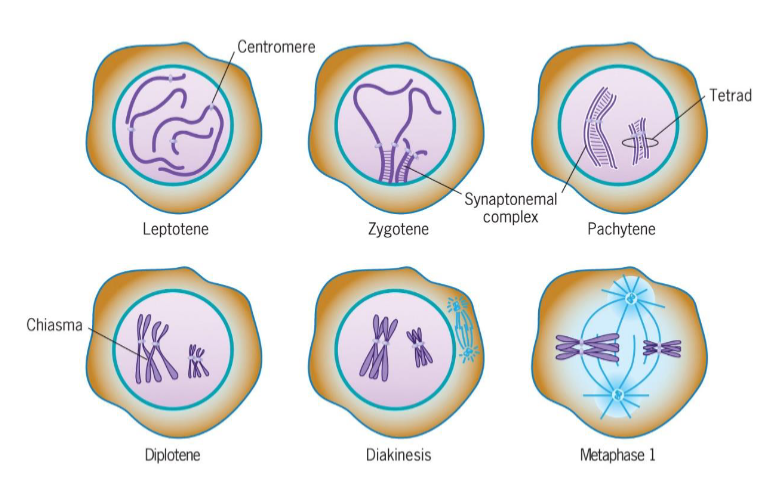
Pachytene (Prophase 1 of meiosis)
synapsis ends, synapsed chromosomes formed tetrads
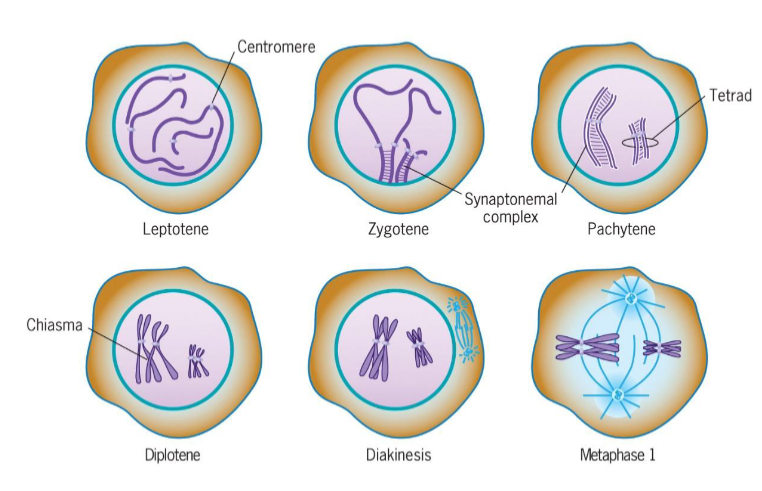
Diplotene (Prophase 1 of meiosis)
Chiasmata occur (crossing over); point of genetic recombination
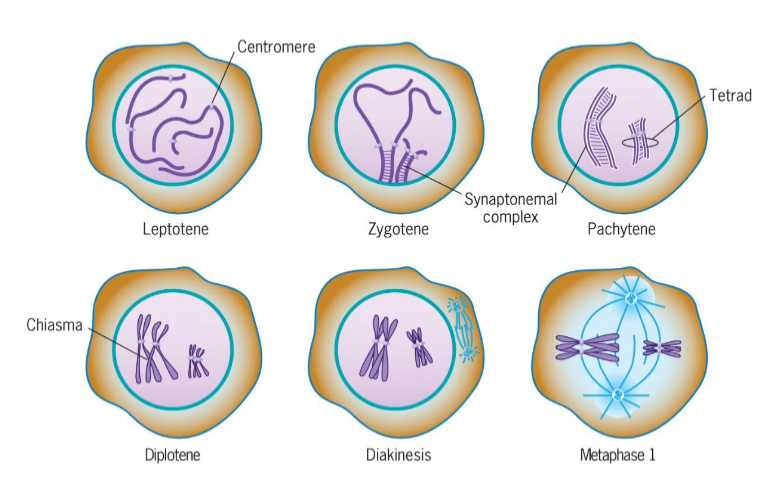
Diakinesis (Prophase 1 of meiosis)
chromosomes prepared for attachment to spindle fibers
ends with disappearance of nucleolus and disassembly of nuclear envelope
triggered by increase in MPF activity
Metaphase I (meiosis)
2 homologous chromosomes aligned at metaphase plate
homologous chromosomes held by one or several chiasmata
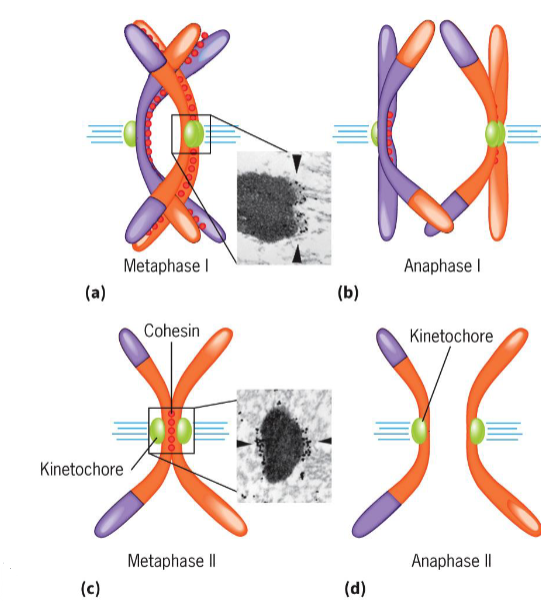
Anaphase I (meiosis)
homologous chromosomes separate
maternal and paternal chromosomes of each tetrad segregate into 2 daughter cells independent of other chromosomes
Telophase I (meiosis)
produces less dramatic changes than telophase of mitosis
nuclear envelope may or may not reform
interkinesis: stage between 2 divisions
Cells communicate with one another through….
extracellular messenger molecules
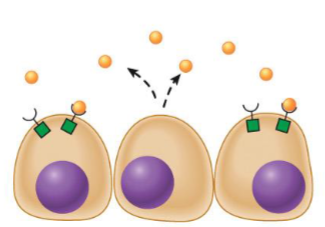
Autocrine messengers
cell has receptors on its surface that respond to messenger
cells releasing message will stimulate/inhibit themselves
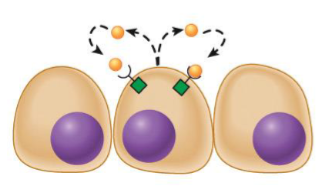
Paracrine messengers
travel short distances through extracellular space
limited in their ability to travel around body because they are inherently unstable, or they are degraded by enzymes, or they bind to extracellular matrix
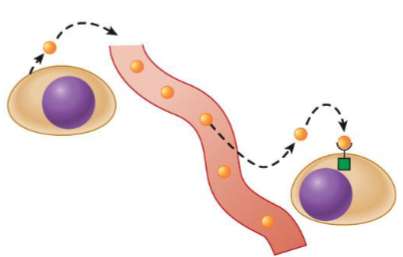
Endocrine messengers (hormones)
reach target cells through bloodstream
also called hormone; act on target cells located at distant sites in body
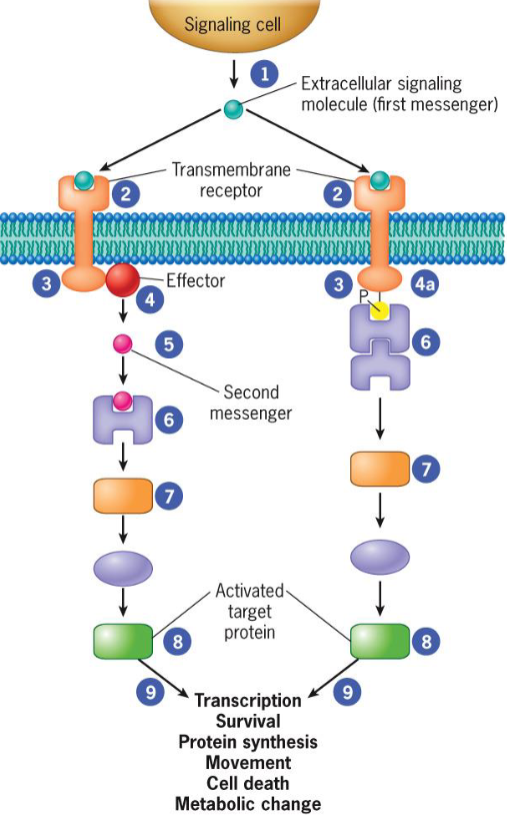
First messenger/Ligand
molecule that binds to receptor
activate receptors that stimulate effectors to give rise to a physiological response
Receptors
in target cells, receive an extracellular message
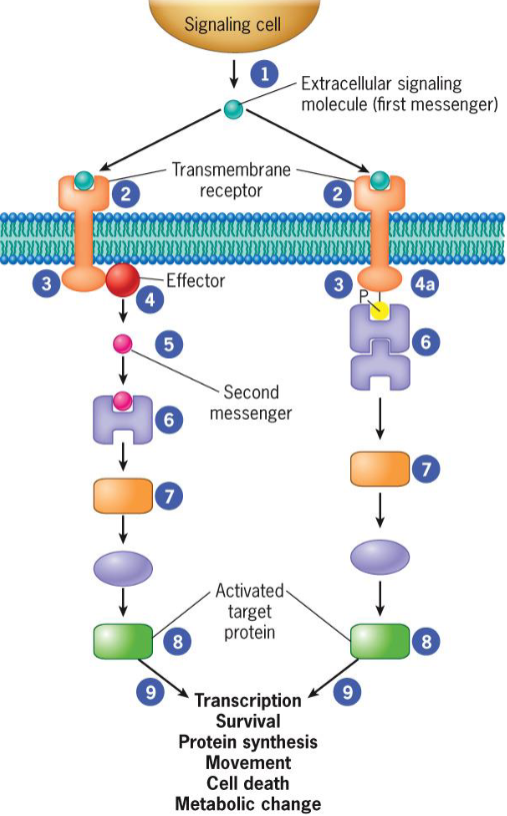
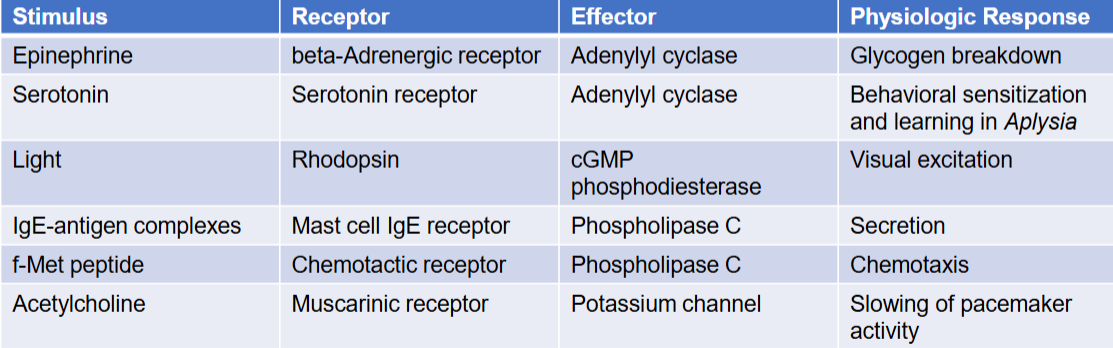
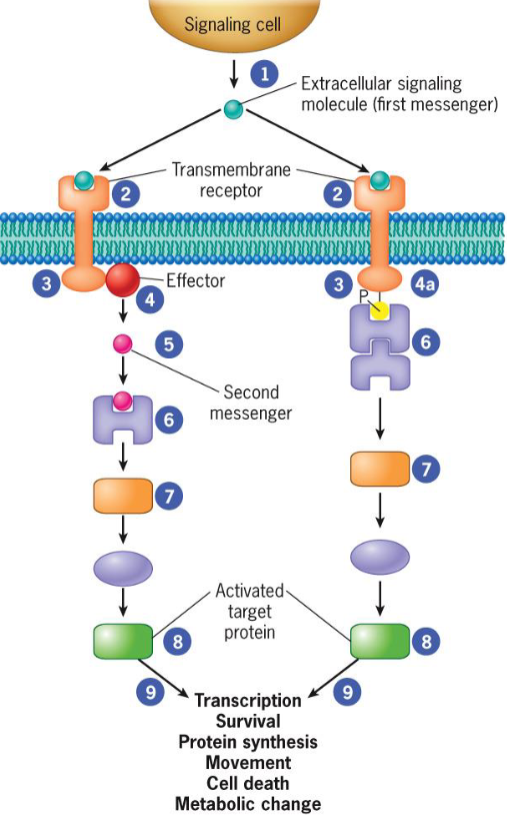
2 different types of signal transduction pathways
activation by diffusible second messenger
recruitment of cytoplasmic proteins to plasma membrane
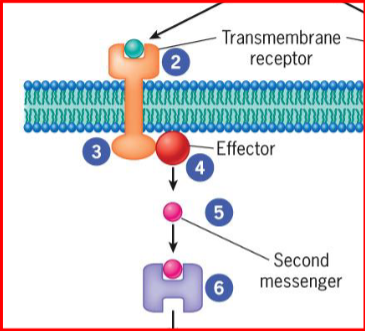
effectors
generated when receptor transmits signal from cytoplasmic domain to nearby enzyme
second messenger/enzyme
small molecules that act as activators/inhibitors of specific proteins
diffuses through cytosol or remains embedded in lipid bilayer of membrane
recruiting station
a transformed cytoplasmic domain of a receptor that allows it to transmit it’s signal for cellular signaling proteins
proteins interact with one another, or with components of cellular membrane

Signaling pathways
a series of proteins
each protein in pathway alters conformation of next protein
protein conformation usually altered by phosphorylation
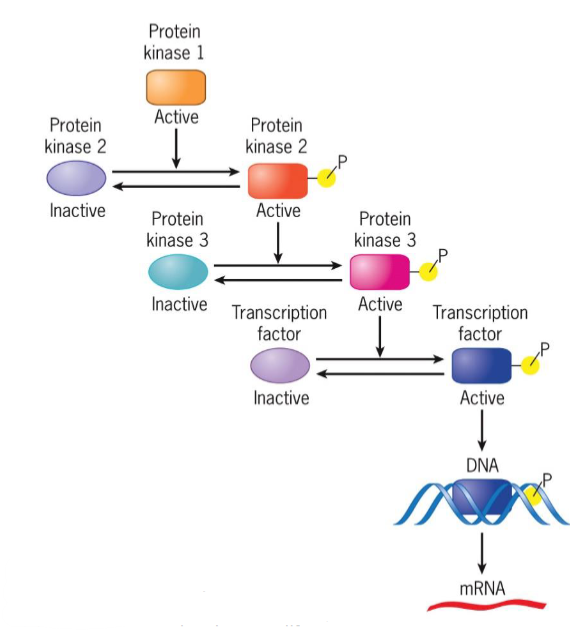
kinases vs phosphatases
kinases add phosphate groups while phosphatases remove them
human genome encodes >500 protein kinases and 150 protein phosphatases
transfer phosphate groups to serine/threonine residues of their protein substrates or phosphorylates tyrosine residues
soluble cytoplasmic proteins or integral membrane proteins.
Protein phosphorylation
can change protein behavior in different ways
activating or inactivating an enzyme
increasing or decreasing protein-protein interactions
changing subcellular location of protein
triggering protein degradation
signal transduction
transmitted signals reach target proteins that ultimately receive message to alter cell activity
changes in cellular activities, gene expression, and ion permeability
alteration of activity of metabolic enzymes
reconfiguration of cytoskeleton
increase/decrease in cell mobility
activation of DNA synthesis
cell death
5 extracellular messengers
small molecules, AAs + derivatives
glutamate, glycine, acetylcholine, epinephrine, dopamine, and thyroid hormone act as neurotransmitters and hormones
Gases such as NO and CO
Steroids, derived from cholesterol
Eicosanoids
peptides and proteins
Steroid hormones (extracellular messenger)
regulates sexual differentiation, pregnancy, carb metabolism, and excretion of sodium and potassium ions
Eicosanoids
nonpolar molecules derived from FA, arachidonic acid
regulate variety of processes including pain, inflammation, blood pressure, and blood clotting.
5 types of receptor types
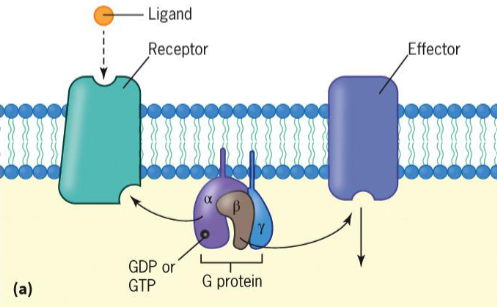
G-protein coupled receptors (GPCRs)
Receptor protein-tyrosine kinases (RTKs)
Ligand gated channels
Steroid hormone receptors
Specific receptors
GPCR
largest superfamily of proteins encoded by animal genomes (1000s)
7 α-helical transmembrane domains
interact with heterotrimeric G Proteins
capable of binding diverse array of ligands
receptor isoforms have different affinities for ligand or interact with different types of G proteins
4 natural ligands that bind to GPCRs
Hormones (both plant and animal)
Neurotransmitters
Opium derivatives
Chemoattractants (odorants, tastants, and photons)
8 steps of Signal Transduction by G Protein-Coupled Receptors
ligand binds to receptor, altering conformation and increasing affinity for G protein to which it binds
Gα subunit releases its GDP, which is replaced by GTP
Gα subunit dissociates from Gβγ complex and binds to effector (adenylyl cyclase), activating effector
activated adenylyl cyclase produces cyclic AMP (cAMP)
Gα reassociates with Gβγ, reforming trimeric G protein, and effector ceases activity.
To prevent overstimulation, receptors must be blocked (desensitization) from continuing to activate G proteins
Receptor is phosphorylated by a G protein-coupled receptor kinase (GRK)
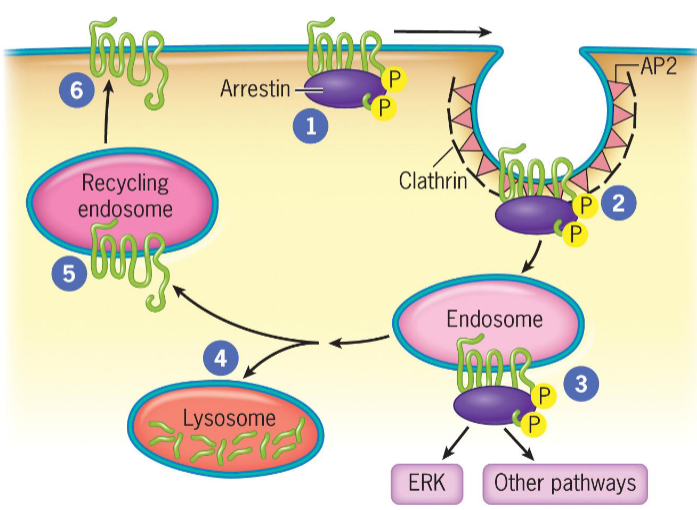
Arrestin molecule binds to phosphorylated receptor to inhibit ligand-bound receptor from activating additional G proteins. Receptor bound to arrestin is likely to be taken up by endocytosis
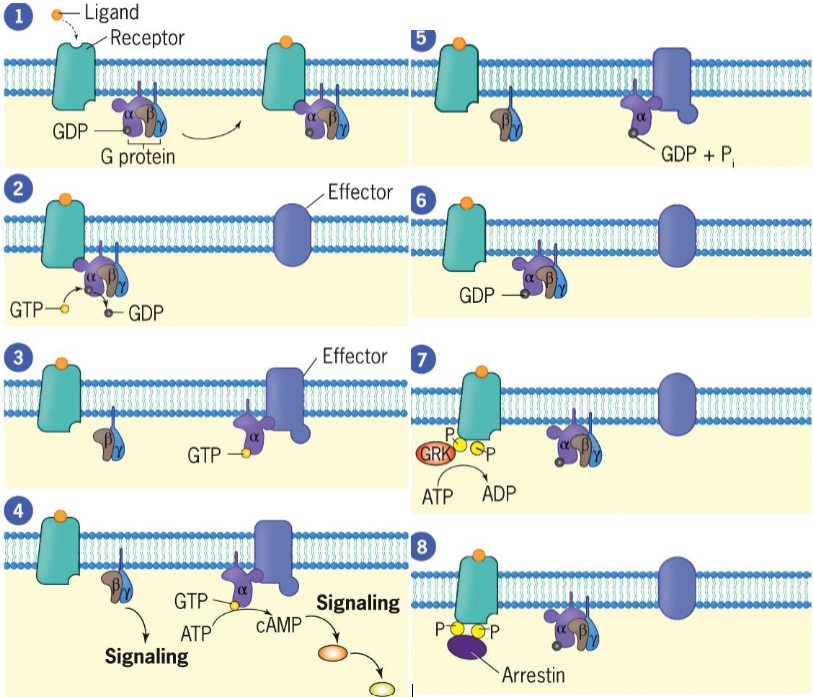
Desensitization
process that blocks active receptors from turning on additional G proteins
Termination of the Response in Signal Transduction by G Protein-Coupled Receptors
Phosphorylation of GPCRs by G protein-coupled receptor kinase allows binding of arrestins (complete for binding with G proteins)
Upon arrestin binding, GPCRs become desensitized
If receptors are recycled and returned to cell surface, cells remain sensitive to ligand and are resensitized
Arrestin-bound GPCRs internalized by clathrin-coated pits that bud into cytoplasm
Clathrin-coated vesicles deliver their contents, including GPCRs, to endosomes.
In endosomes, arrestins are scaffolds for assembly of signaling complexes (MAPK cascade and transcription factor ERK)
GPCRs delivered to lysosomes for degradation OR returned to plasma membrane in recycling endosome, where they interact with new extracellular ligands