AP Biology Review May 2025
1/106
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
107 Terms
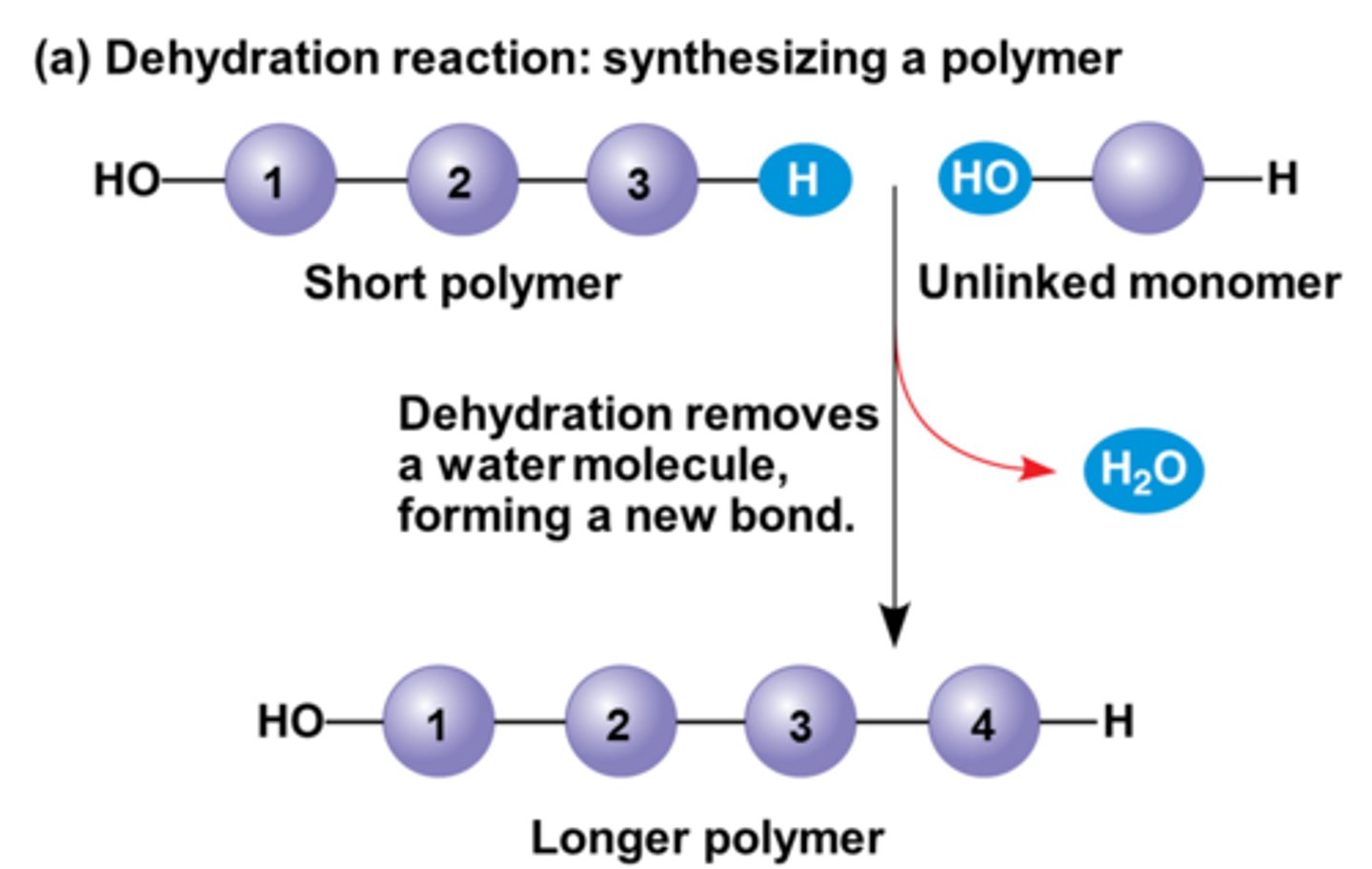
Dehydration
connecting monomers together by the removal of water
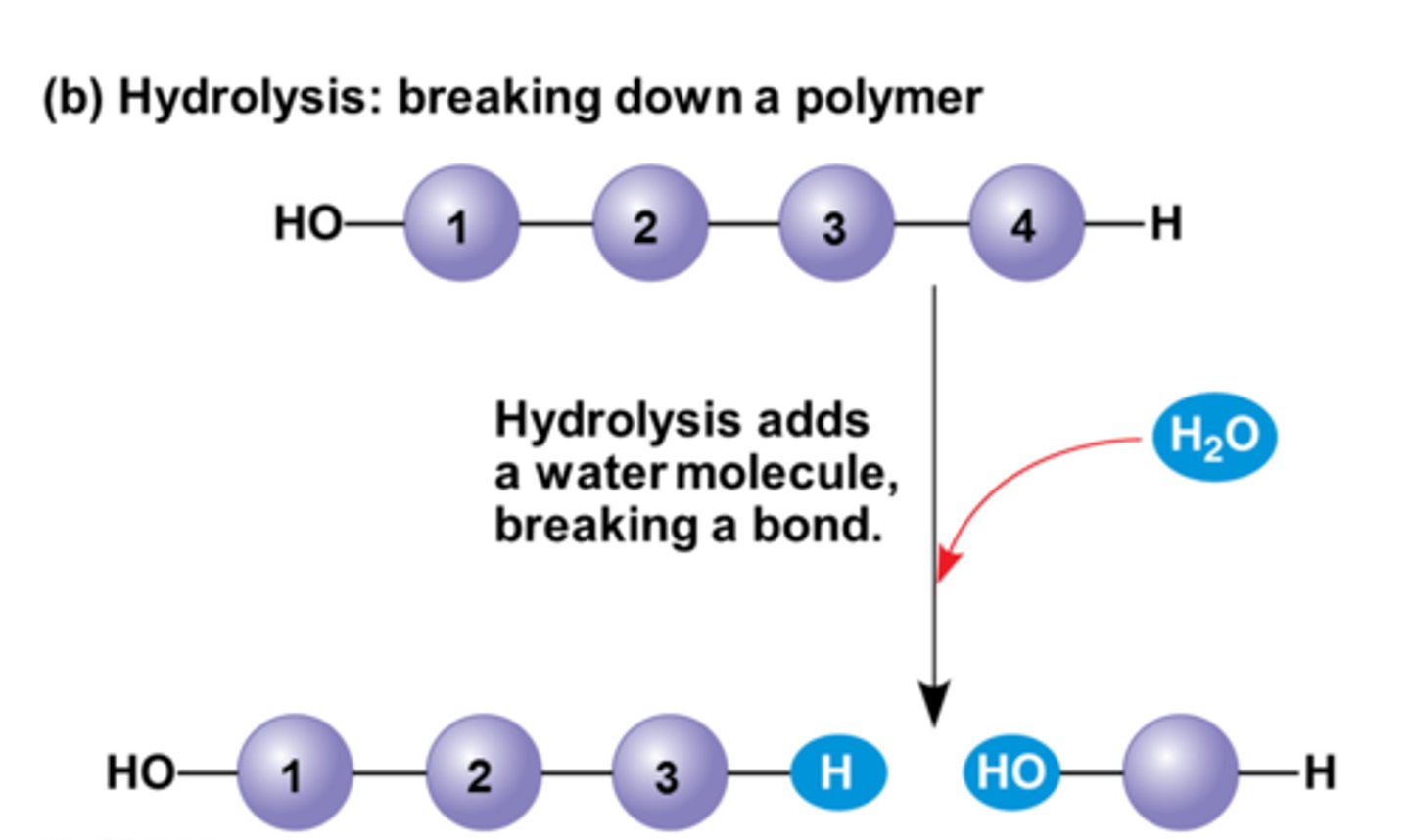
Hydrolysis
disassembling polymers by the addition of water
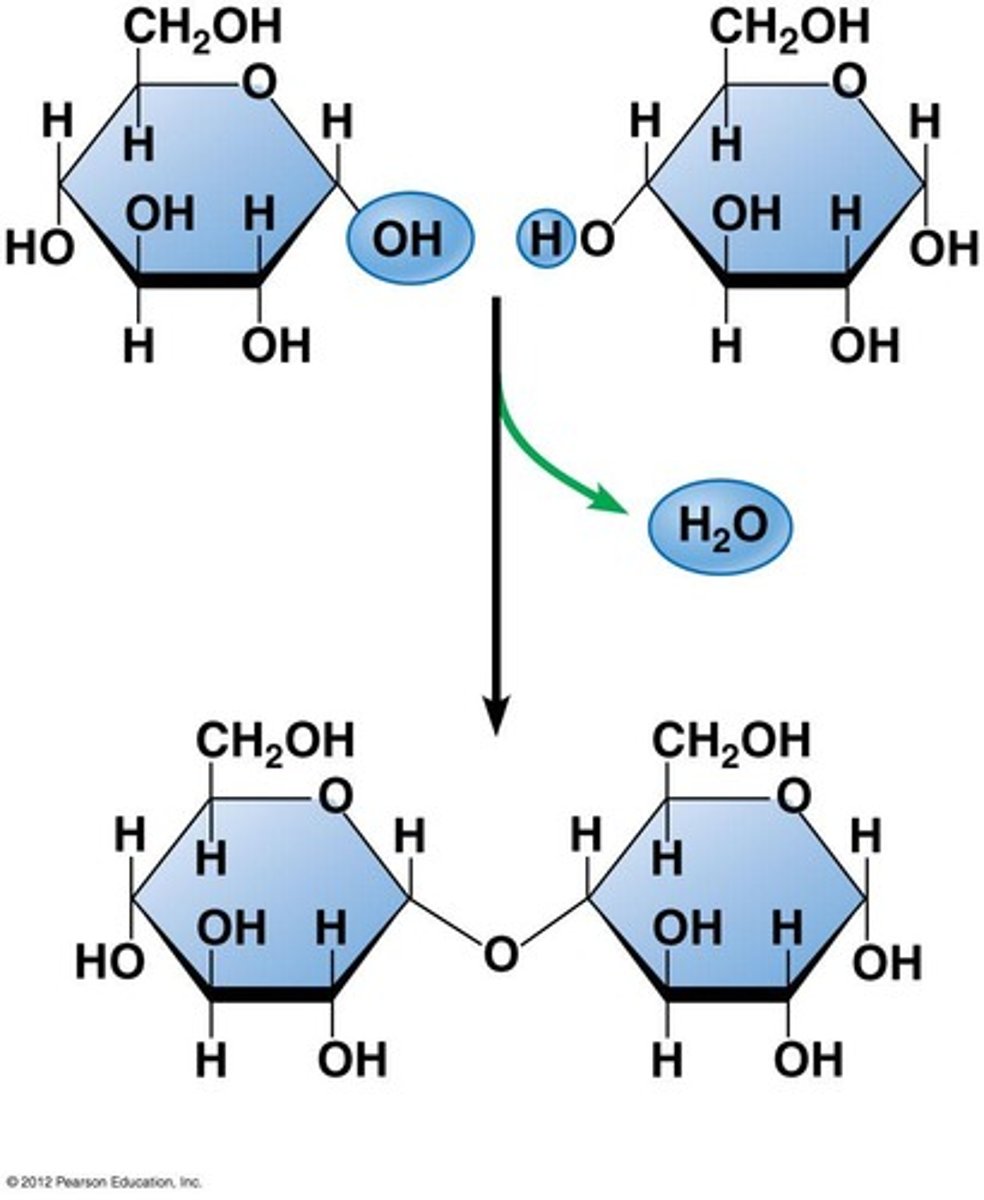
Disaccharides
glucose + glucose = maltose / glucose + fructose = sucrose / glucose + galactose = lactose
Polysaccharides
Plants: starch (energy) and cellulose (structure)
Animals: glycogen (energy) and chitin (structure)
*Lipids
hydrophobic (very non-polar), consist of long hydrocarbon chains
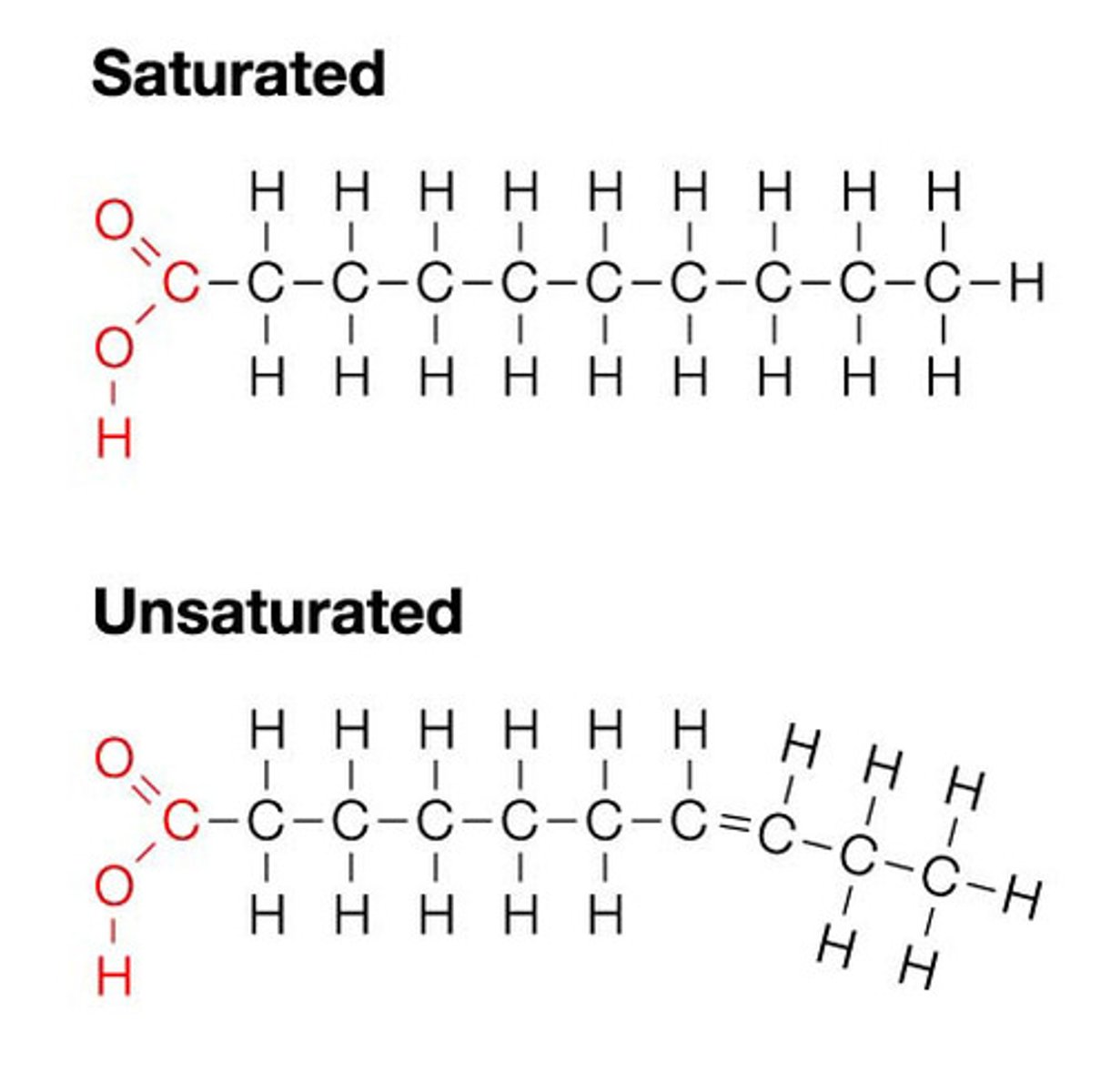
Fats
consist of glycerol and 3 fatty acids, store long term energy, saturated = no double bond in hydrocarbon tails (no kink), unsaturated = double bond (kink)
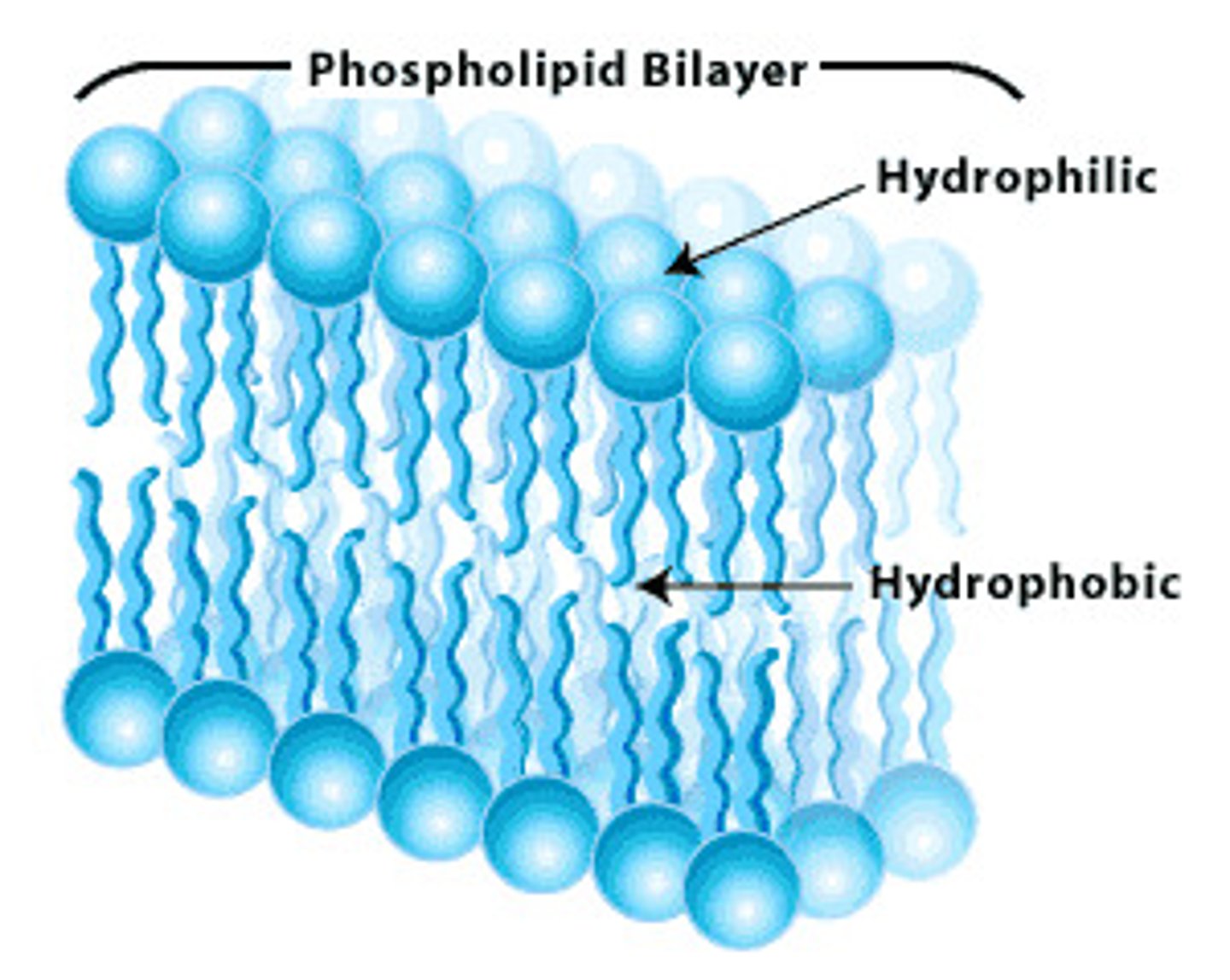
Phospholipids
consist of phosphate head, glycerol, and 2 fatty acid tails, tail is hydrophobic, head is hydrophillic
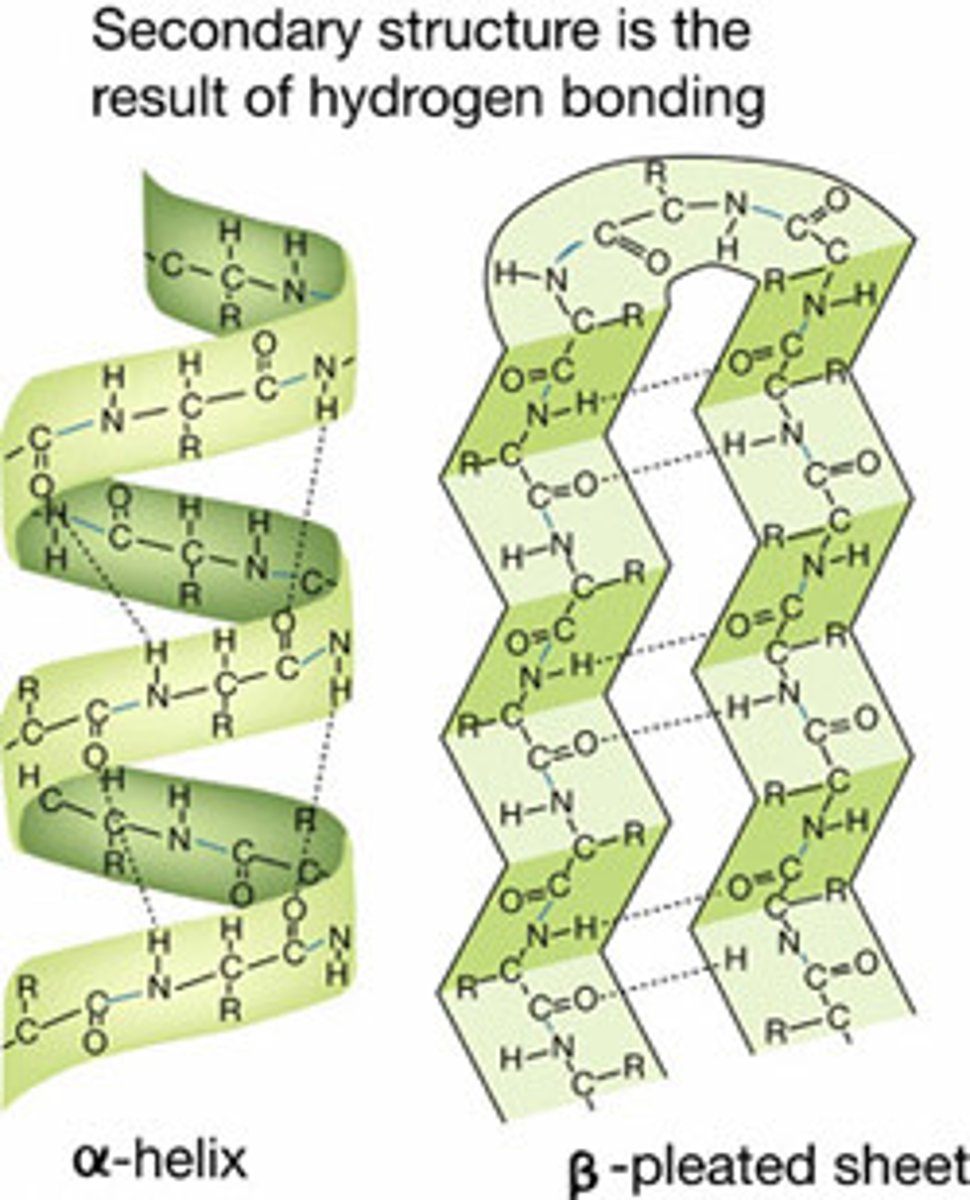
Protein structure and organization
composed of an amino group, a carboxyl group, hydrogen, and an R group, joined by peptide bonds and folded numerous times; 1) Primary (linear sequence) 2) Secondary (helix or pleat) 3) Tertiary 4) Quaternary (globular)
Protein functions (8)
1) enzymes 2) antibodies 3) storage proteins 4) transport proteins 5) hormones 6) receptor proteins 7) motor proteins 8) structural proteins
*Nucleic Acids
DNA (A+T, G+C) carries genetic info, RNA (A+U, G+C) manufactures proteins
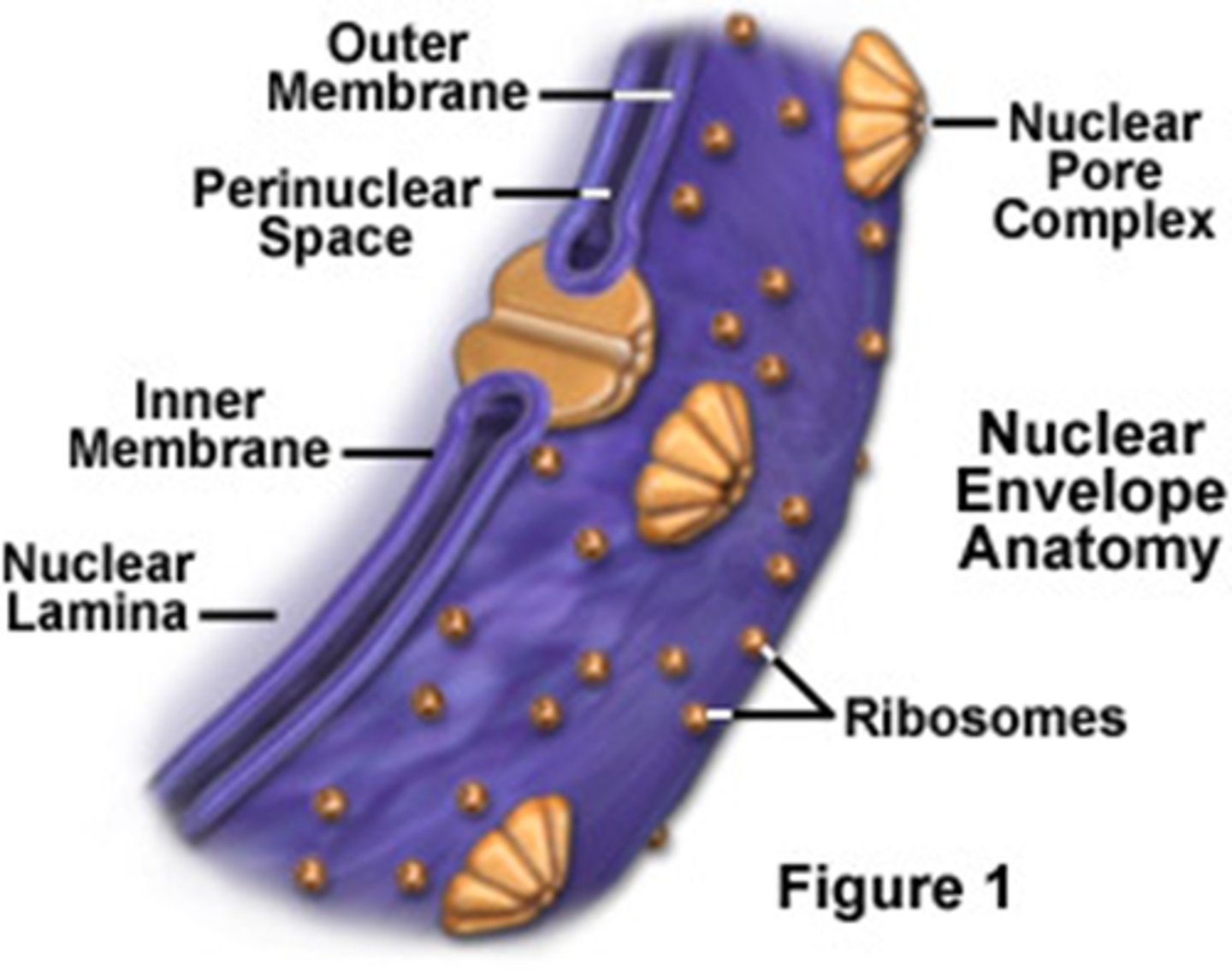
Nuclear Envelope
double membrane enclosing the nucleus (where genetic info is stored) perforated with pores, continuous with ER
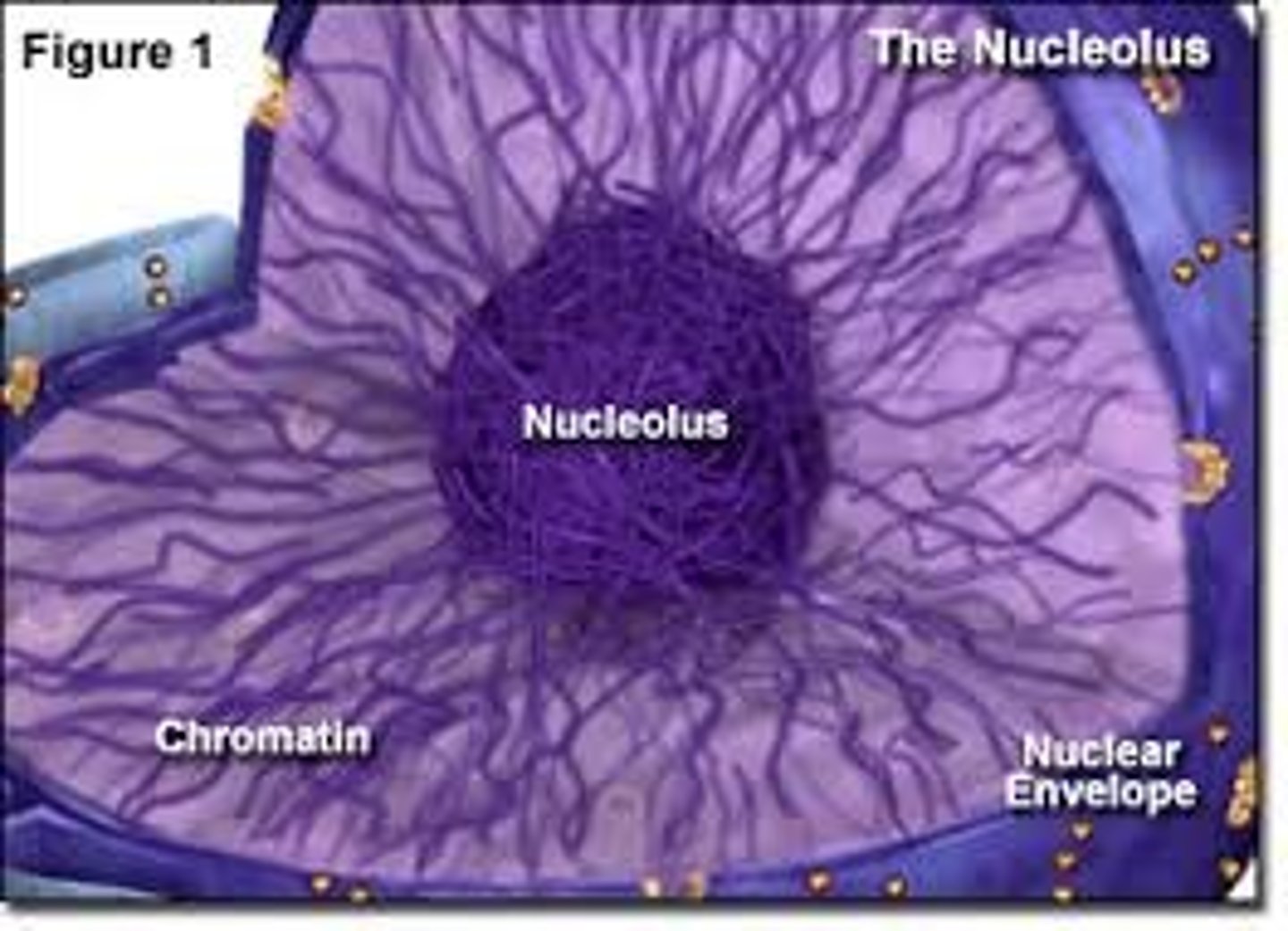
Chromatin
uncondensed DNA that forms chromosomes during cell division
Nucleolus
nonmembranous structure involved in production of ribosomes, a nucleus has one or more of these
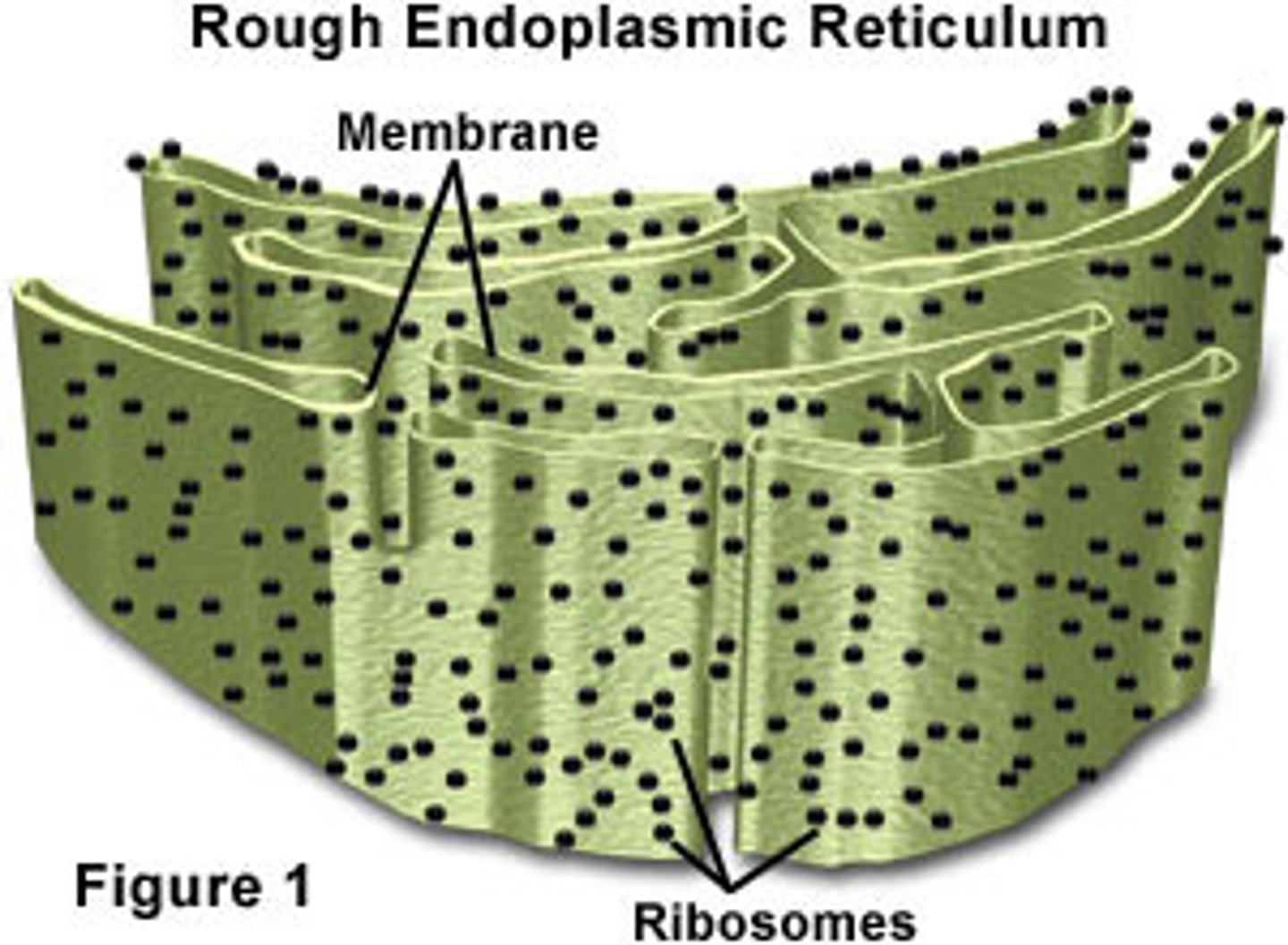
Rough ER
covered in ribosomes, secretes and transports proteins produced by ribosomes
Smooth ER
metabollic processes (synthesis of lipids, metabolism of carbs, detoxification of drugs and poisons)
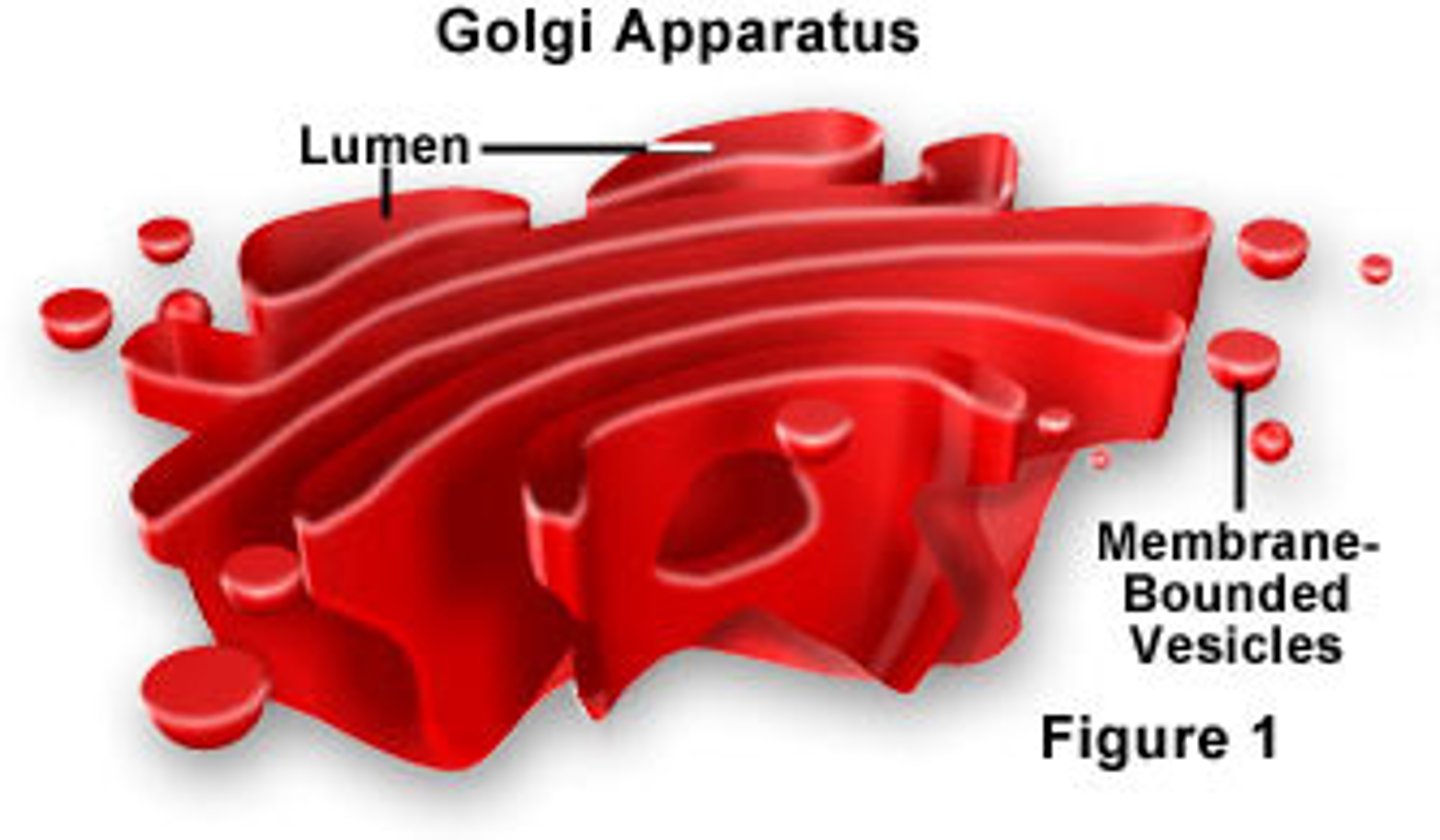
Golgi
stores, transports, and secretes cell products
Cytoskeleton
supports cell, maintains its shape, aids in movement of cell products
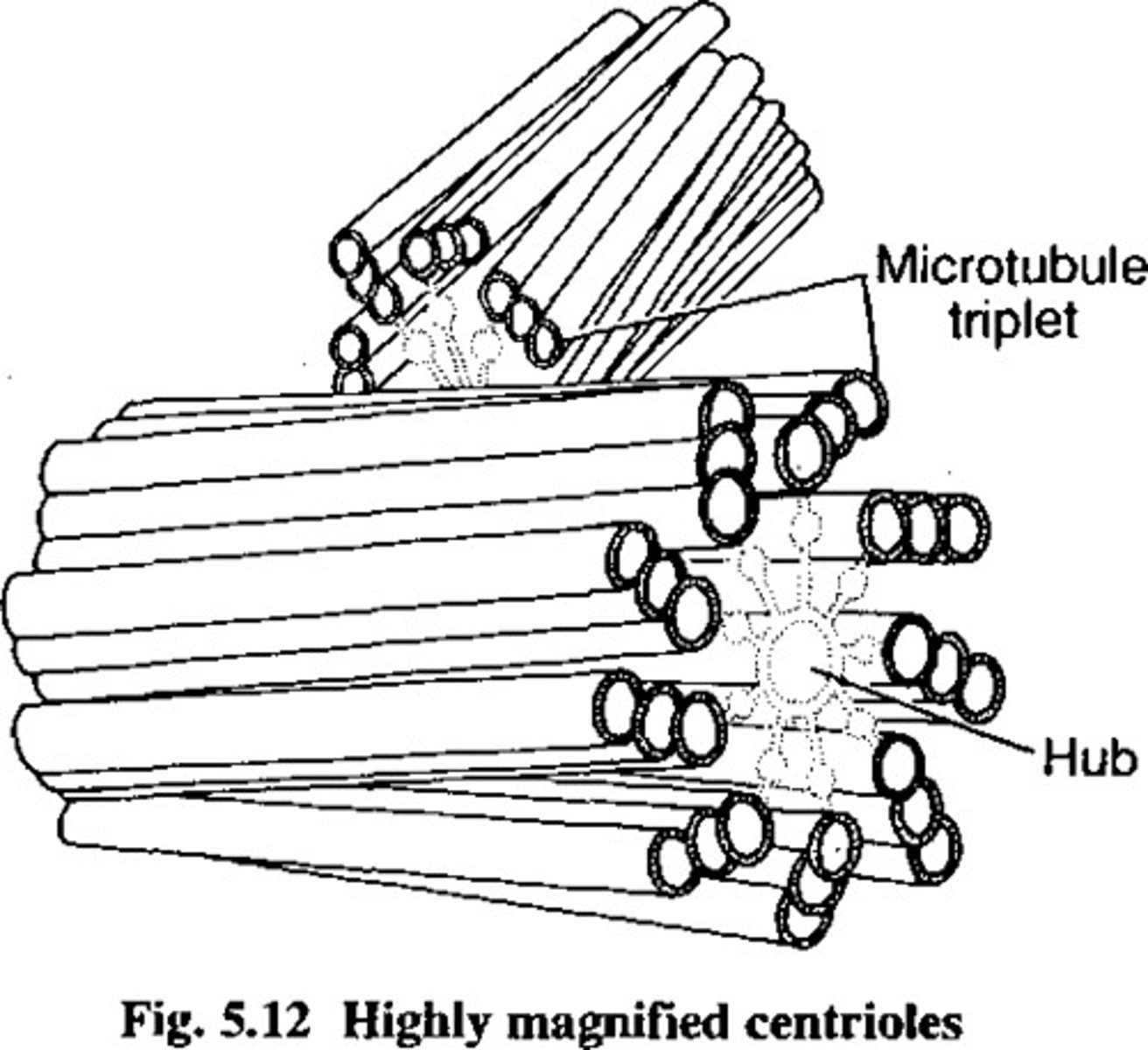
Centrosomes (2 centrioles)
only in animal cells, microtubules used for cell division
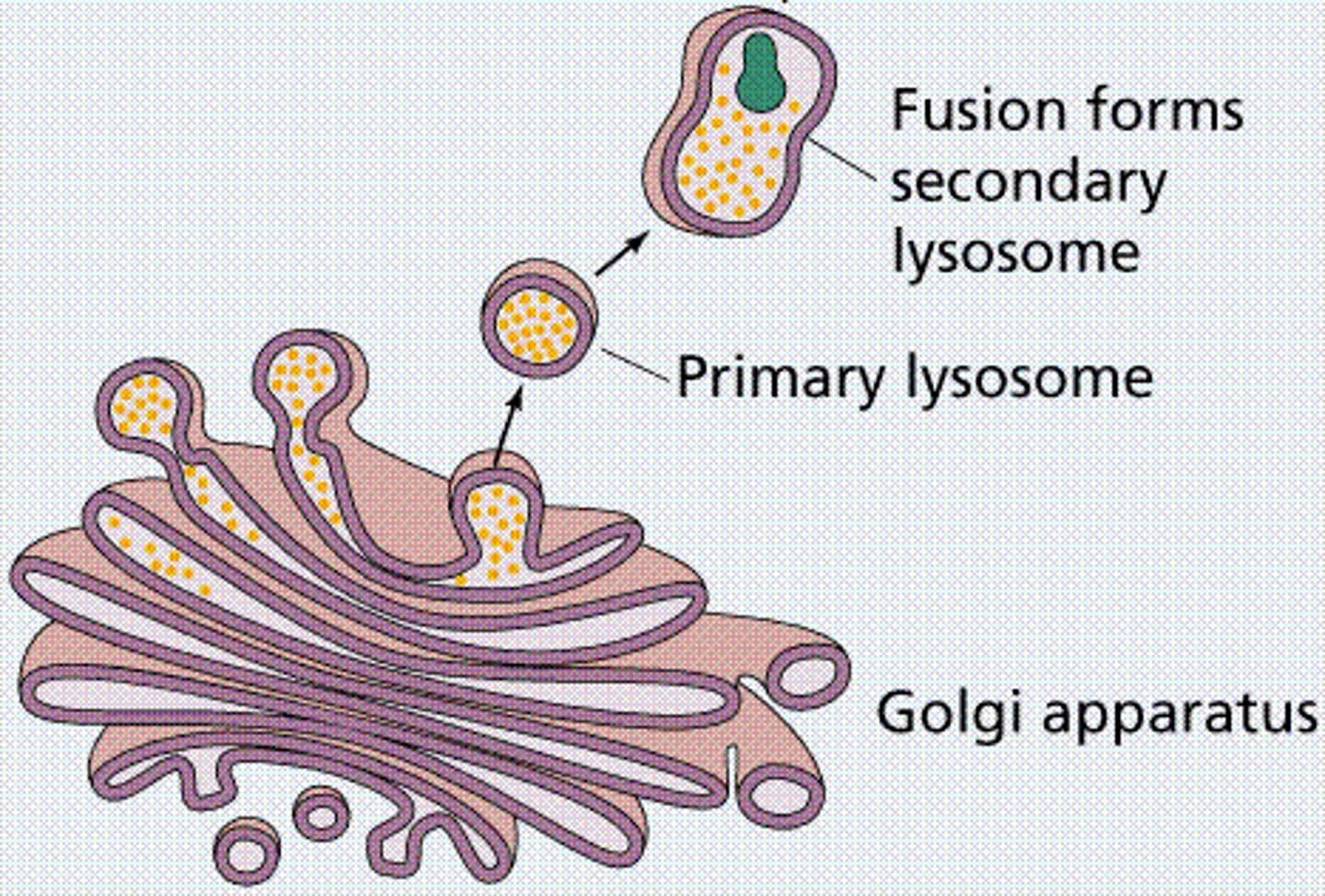
Lysosomes
only in animal cells, digestive organelles
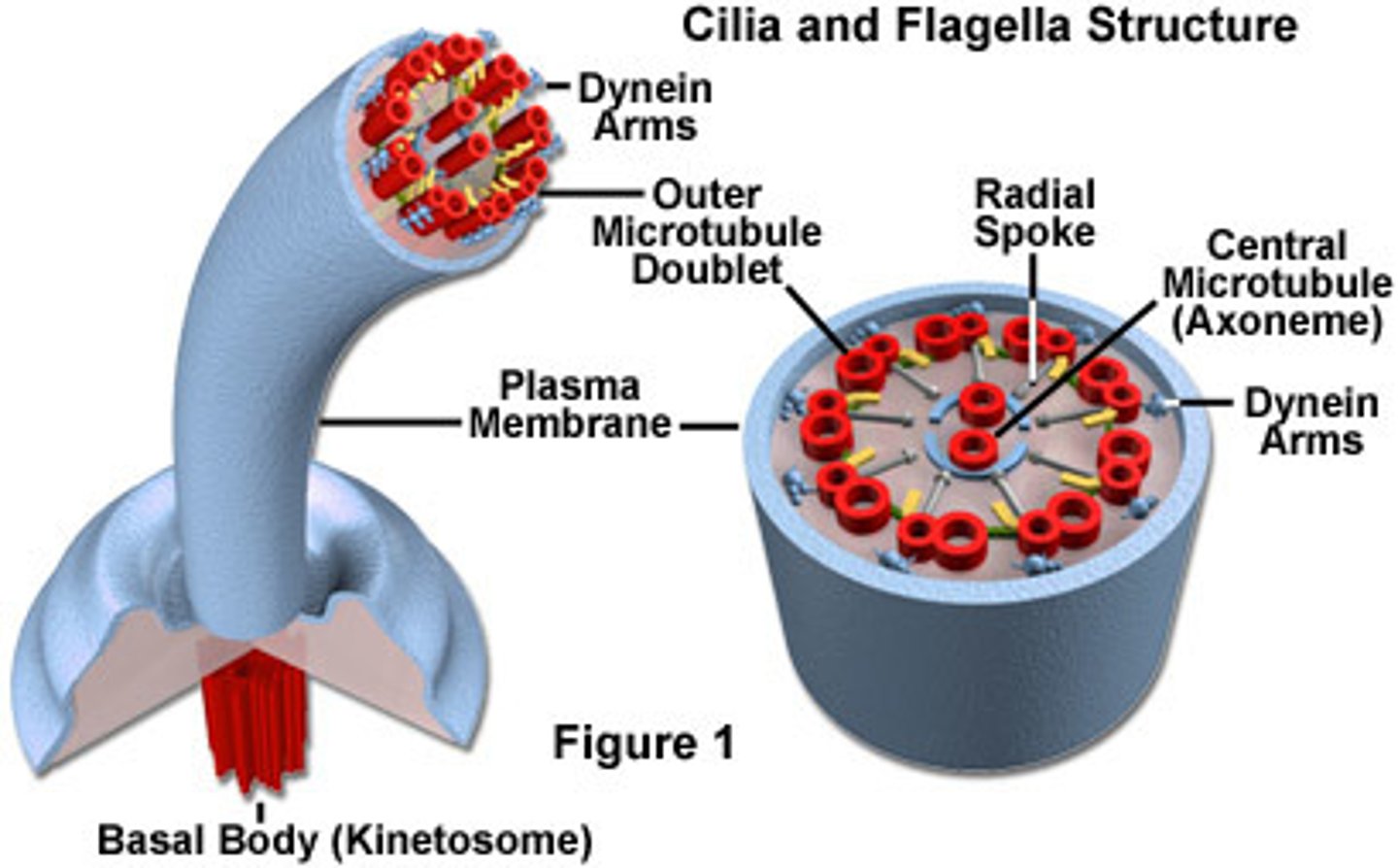
Flagella
only in animal cells, cluster of microtubules for motility
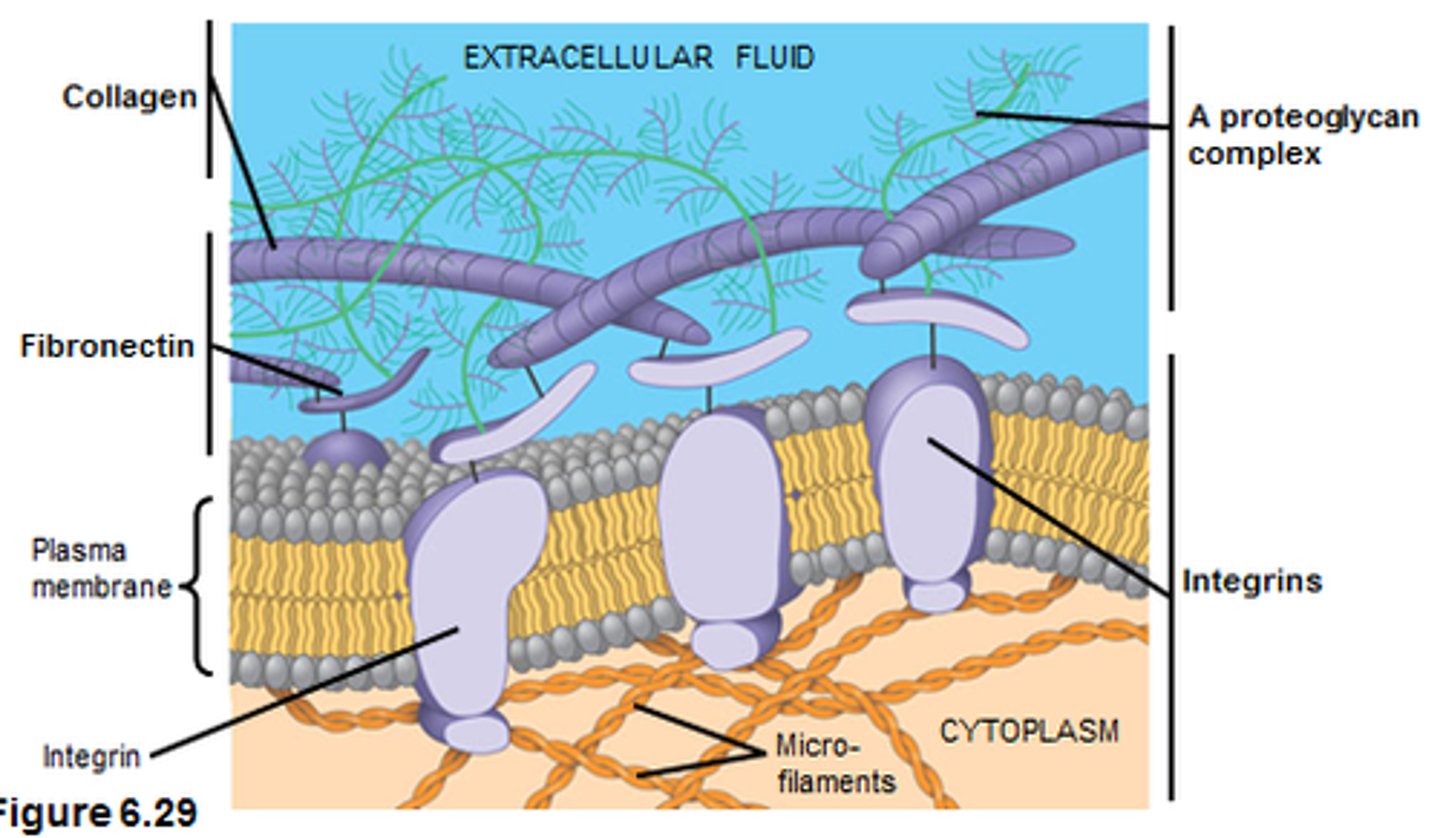
Extracellular Matrix
only in animal cells, made of proteins that provide support for cells and relay information for communication between the environment and the cell
Central Vacuole
only in plant cells, stores water and sugar, breaks down waste, and used as a mechanism for plant growth (when it swells)
Prokaryotic vs. Eukaryotic
nucleoid / nucleus; only ribosomes / complex membrane-bound organelles; both have same genetic coding, sugars, and amino acids
Phospholipid Bilayer
tails of phospholipids are loosely packed and are in constant motion; membrane contains integral and peripheral proteins, cholestrol, and glycopreotins and glycolipids; cholesterol makes the membrane less permeable to water and other substances; non-polar and small polar molecules can pass through unadied
Passive trasport
movement of molecules without requirement of energy: 1) diffusion 2) osmosis (across a membrane) 3) facilitated diffusion (helped by transport proteins)
Active transport
movement of molecules that requires energy: 1) sodium-potassium pumps 2) exocytosis 3) endocytosis (phagocytosis, pinocytosis)
Membrane Potential
voltage across a membrane due to difference in positive and negative ions, electrons move from high to low concentration (ex. sodium-potassium pumps in neurons)
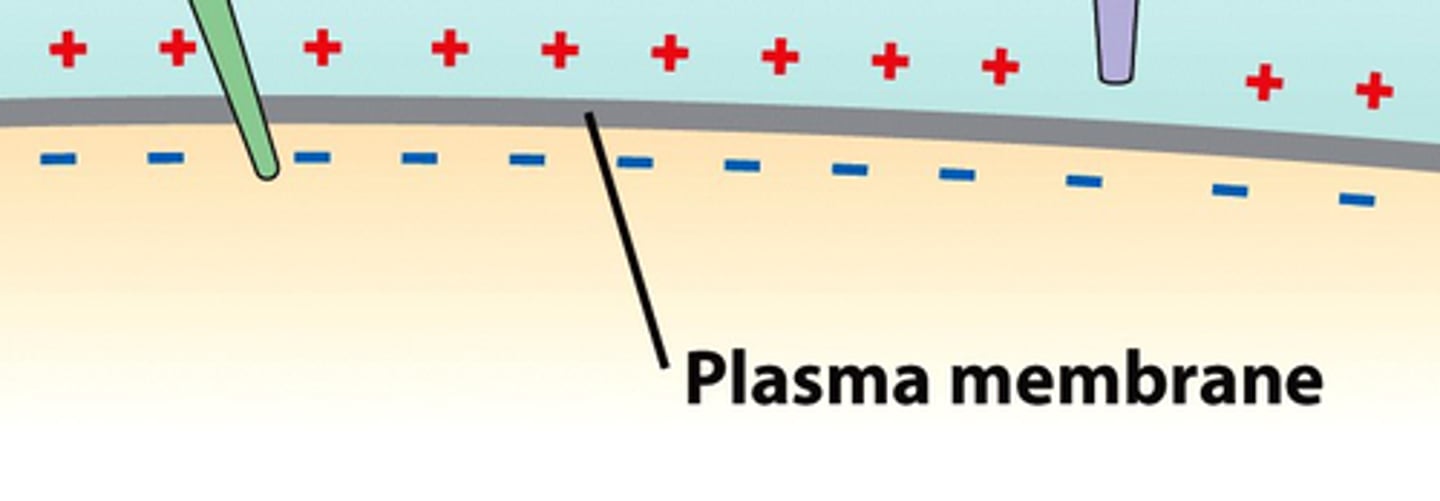
Electrochemical Gradient
diffusion gradient resulting in combination of membrane potential and concentration gradient
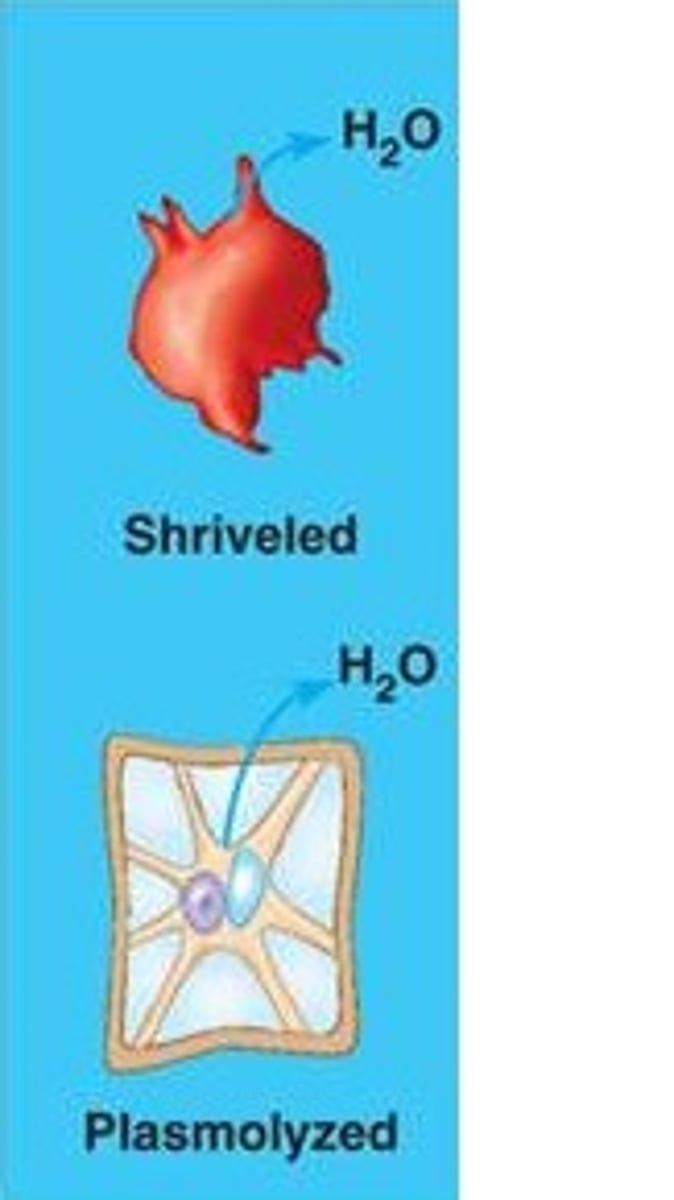
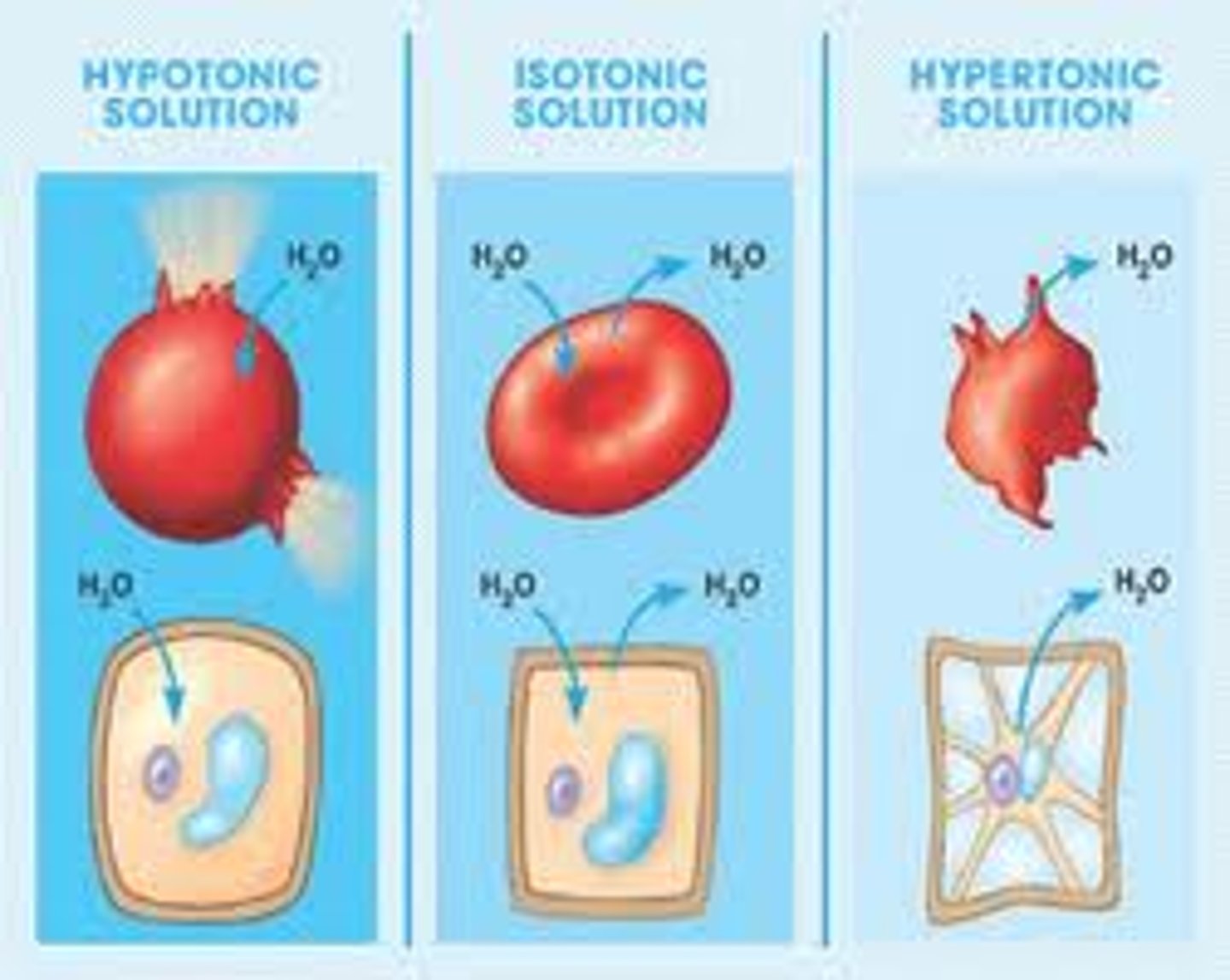
Hypertonic
solution with higher concentration of solutes, animal/plant cell in this solution would become shiveled/plasmolyzed
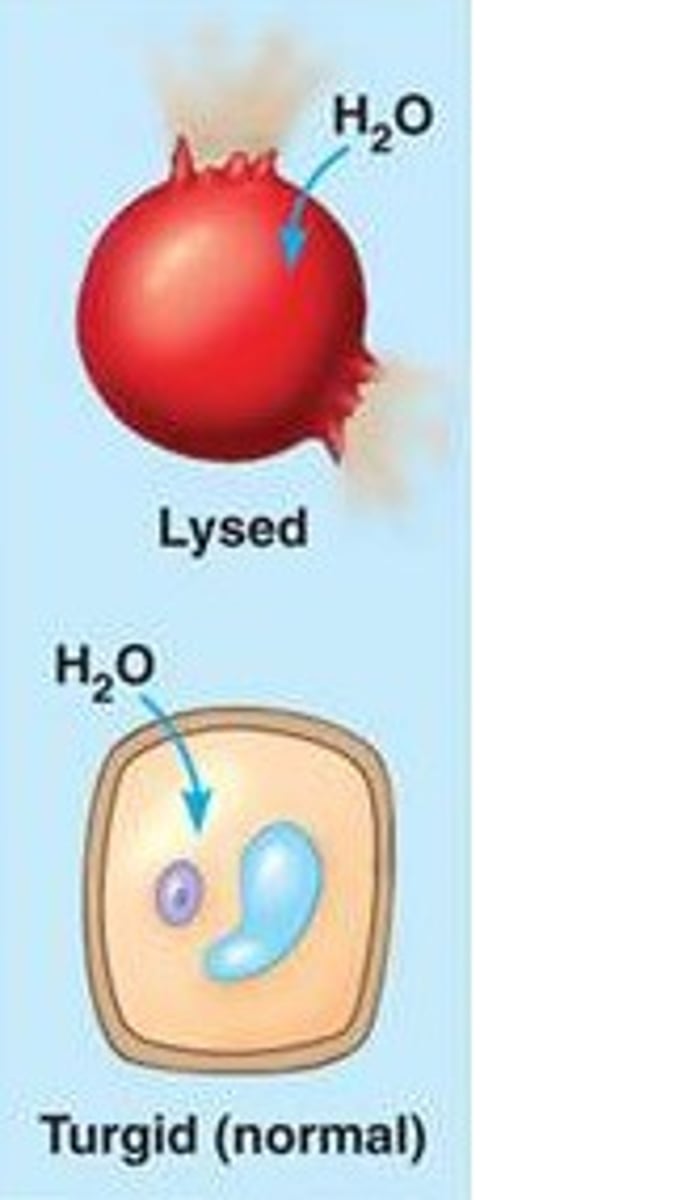
Hypotonic
solution with lower concentration of solutes, animal/plant cell in this solution would lyse/become turgid
Isotonic
equal levels of solute concentration, plant cell in this solution would become flaccid
When ΔG is negative...
...the reaction is exergonic (loss of free energy).
When ΔG is positive...
...the reaction is endergonic (gain of free energy).
*Enzymes
proteins that are biological catalysts, lower the activation energy required to start a chemical reaction (reactants at unstable transition state) can be used over and over
Substrate
the substance that an enzyme acts upon
Active Site
region of enzyme that binds to the substrate
Induced fit
change in the shape of an enzyme's active site induced by the substrate, helps to break down the substrate
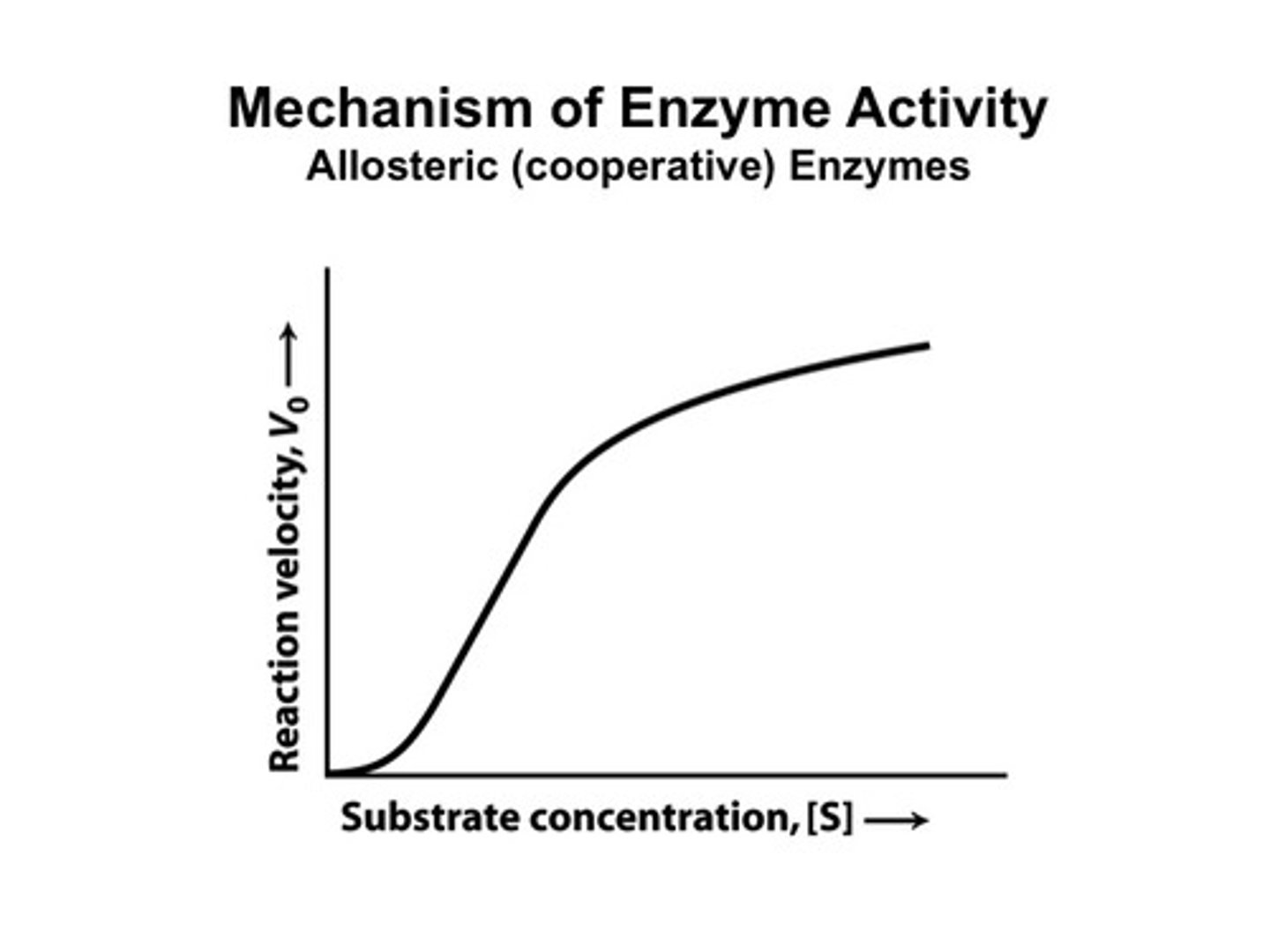
The higher the substrate concentration...
...the faster the reaction until the enzyme becomes saturated.
Denaturation
the unraveling of an enzyme due to high temperatures or incompatible pH
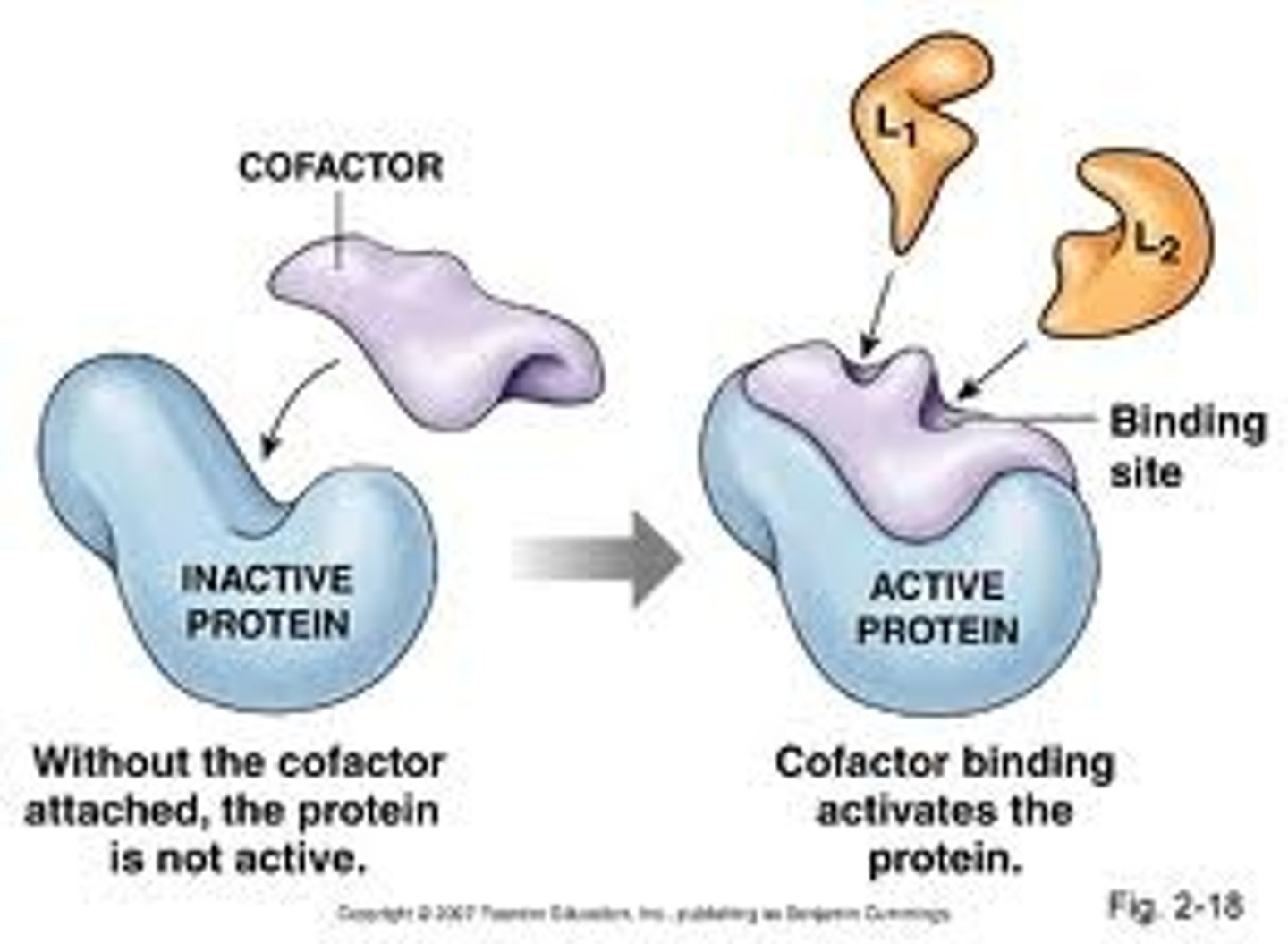
Cofactors
nonprotein molecules that are required for proper enzyme function, cofactors made of organic molecules are called coenzymes
Enzyme inhibition may be irreversible if...
...the inhibitor attaches by covalent bonds (poisons, toxins)
Competitive Inhibitors
resemble a substrate and block enzymes' active sites, can be overcome with higher concentration of substrate

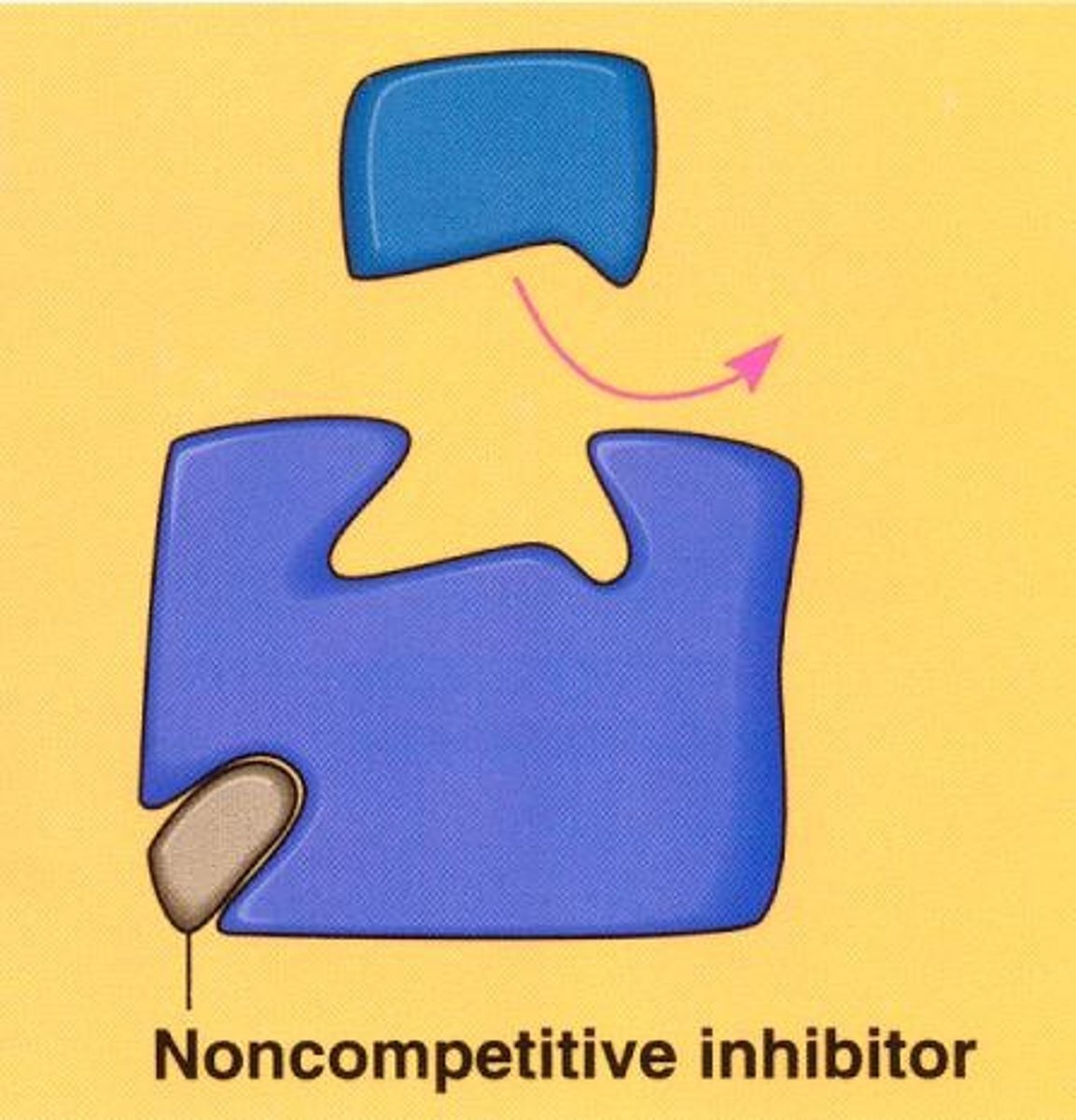
Noncompetitive Inhibitors
bind to a portion of the enzyme and change the shape of the active site so that it cannot match with substrates, used for regulating metabolic reactions
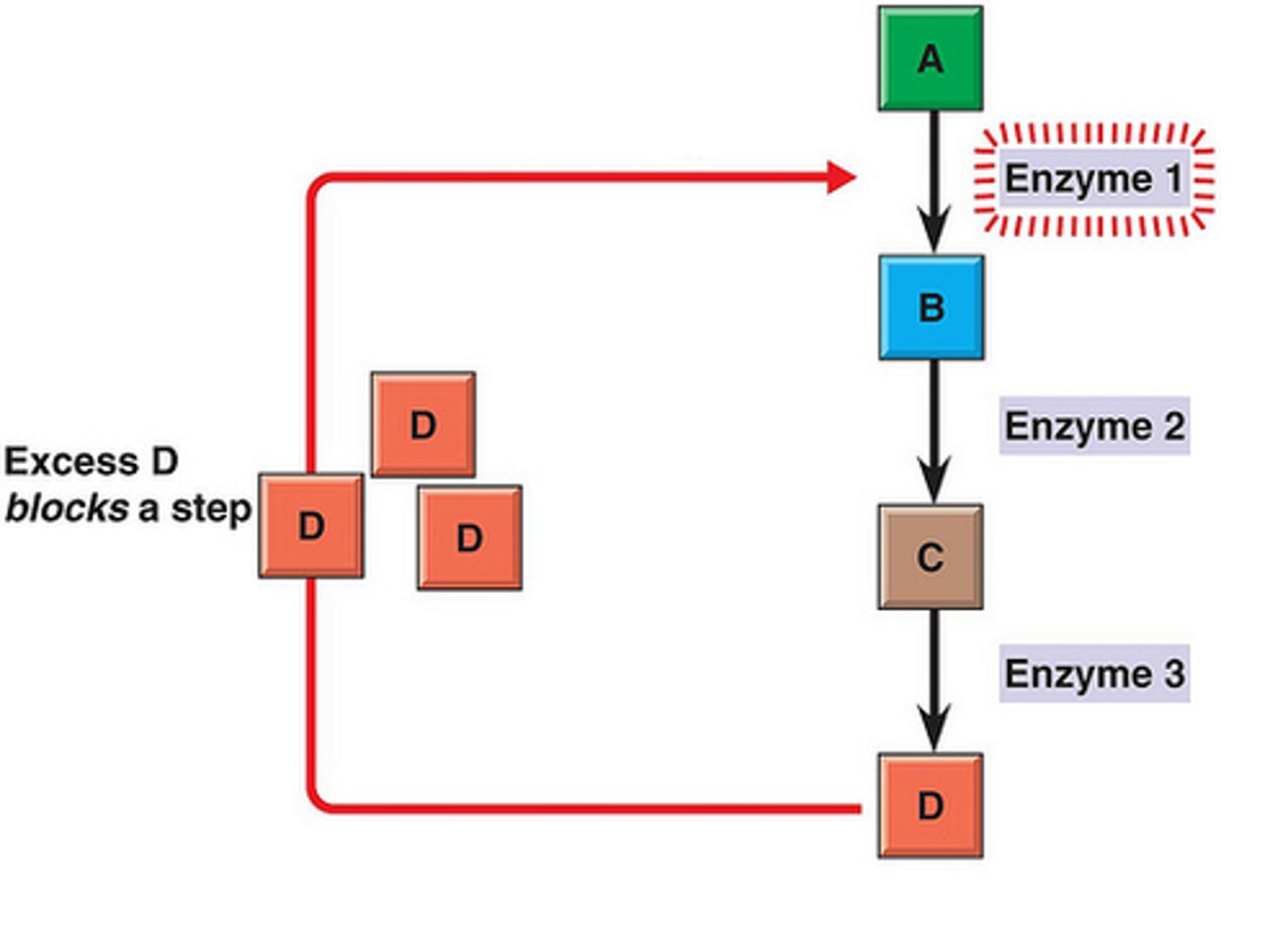
Feedback Inhibition
the product of a metabolic pathway switches off the enzyme that created it earlier in the process
Oxidation
loss of electrons (OIL)
Reduction
gain of electrons (RIG)
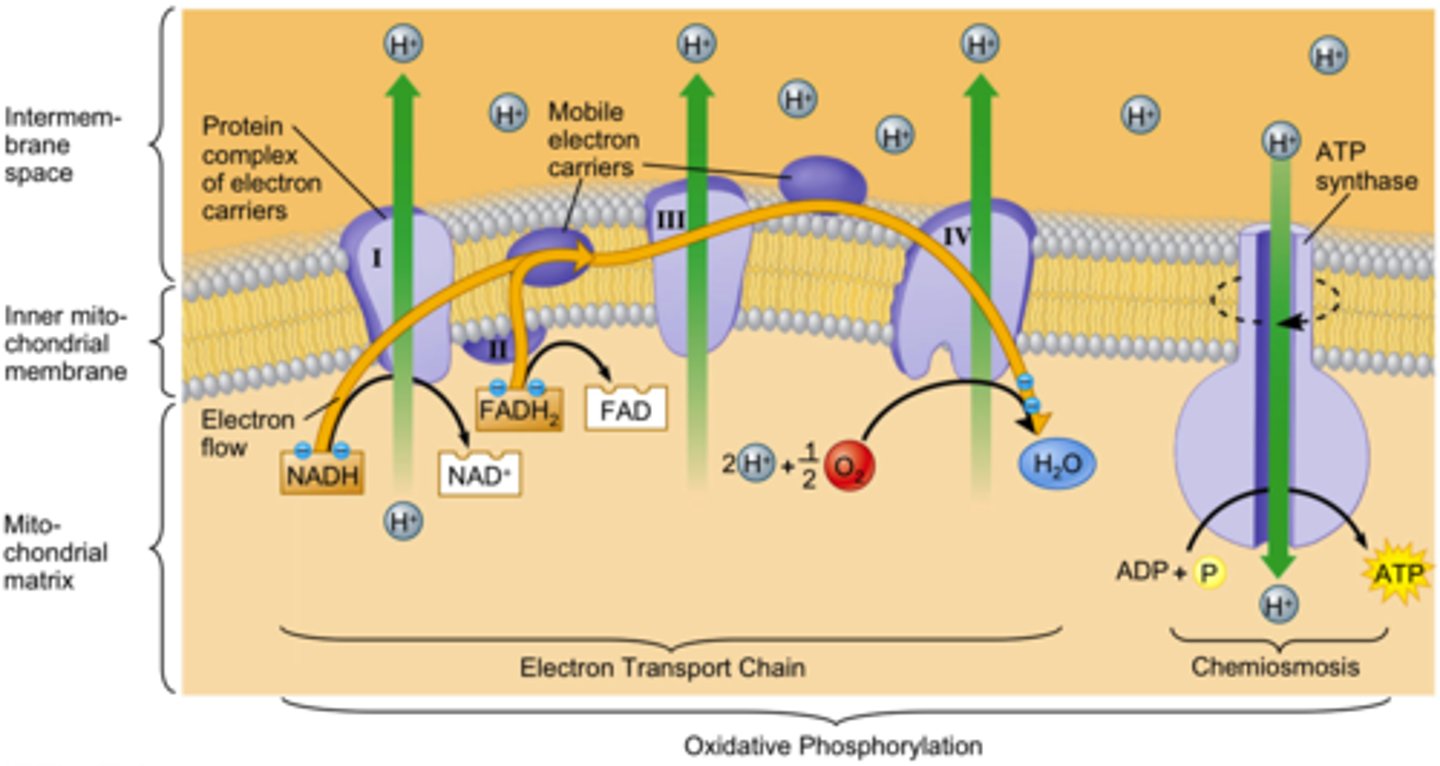
Oxidative Phosphorylation
ATP synthesis powered by redox reactions that transfer electrons to oxygen
Electron Acceptors
Cellular respiration: NAD+ and FAD (to NADH and FADH2)
Photosynthesis: NADP+ (to NADPH)
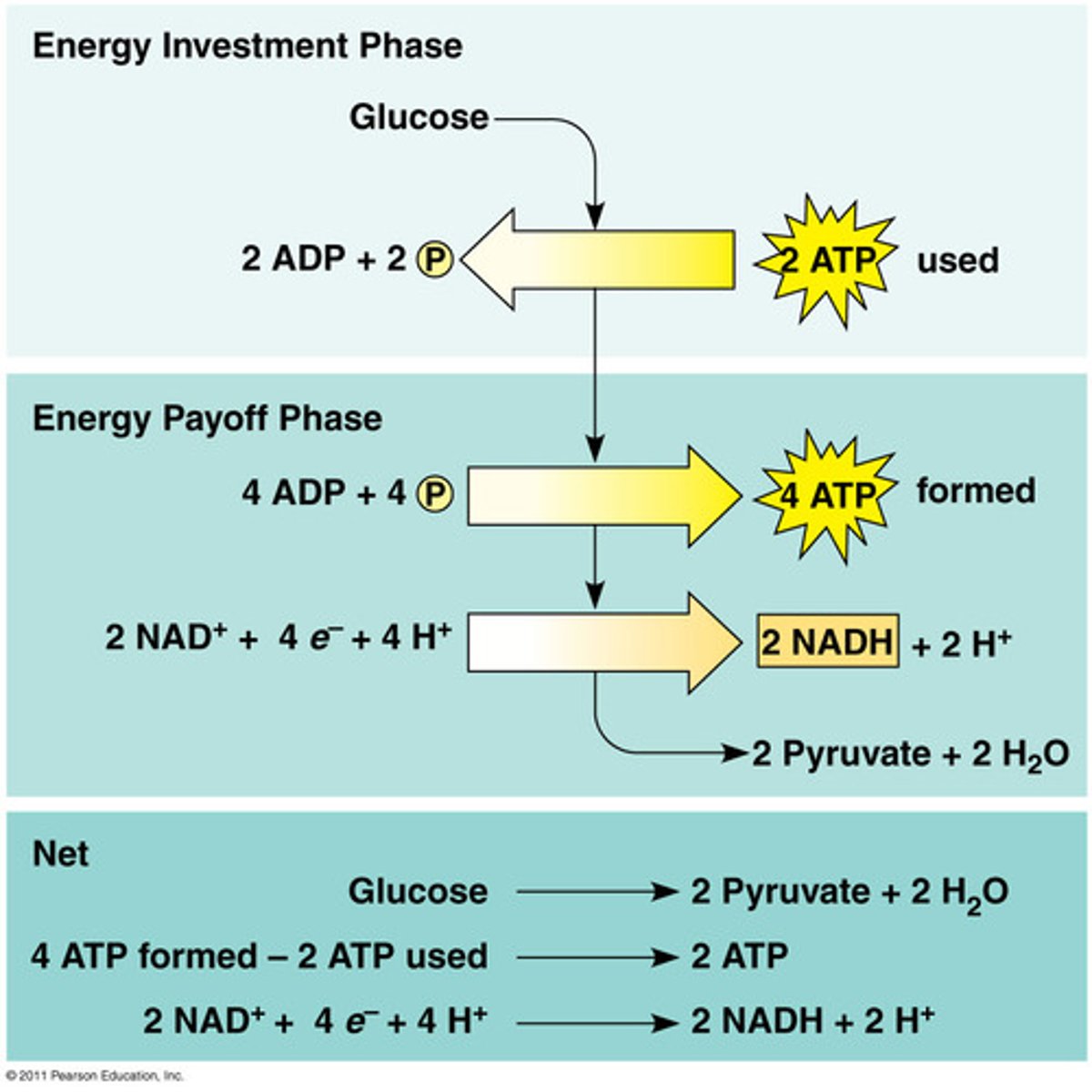
Glycolysis
Input: glucose, 2 ATP
Output: 2 pyruvic acid, 4 ATP (net 2), 2 NADH
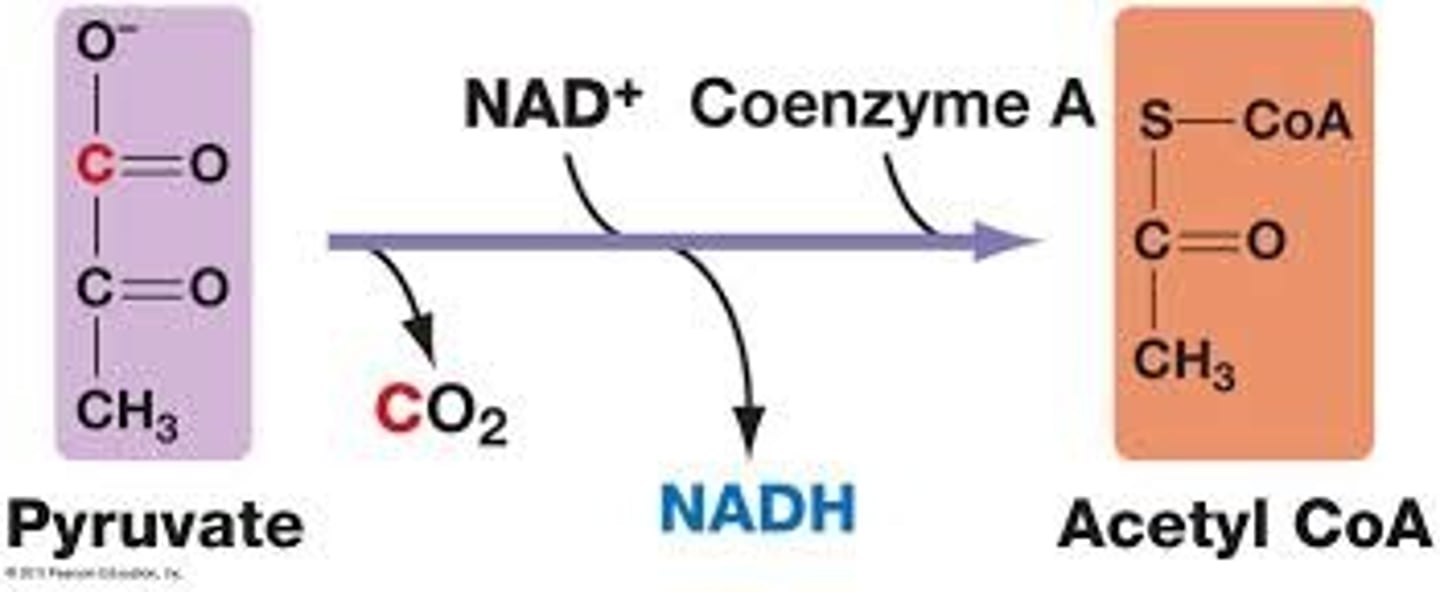
Conversion Reaction before Kreb's
Input: 2 pyruvate
Output: 2 acetyl (w/ CoA), 2 NADH, 2 CO2
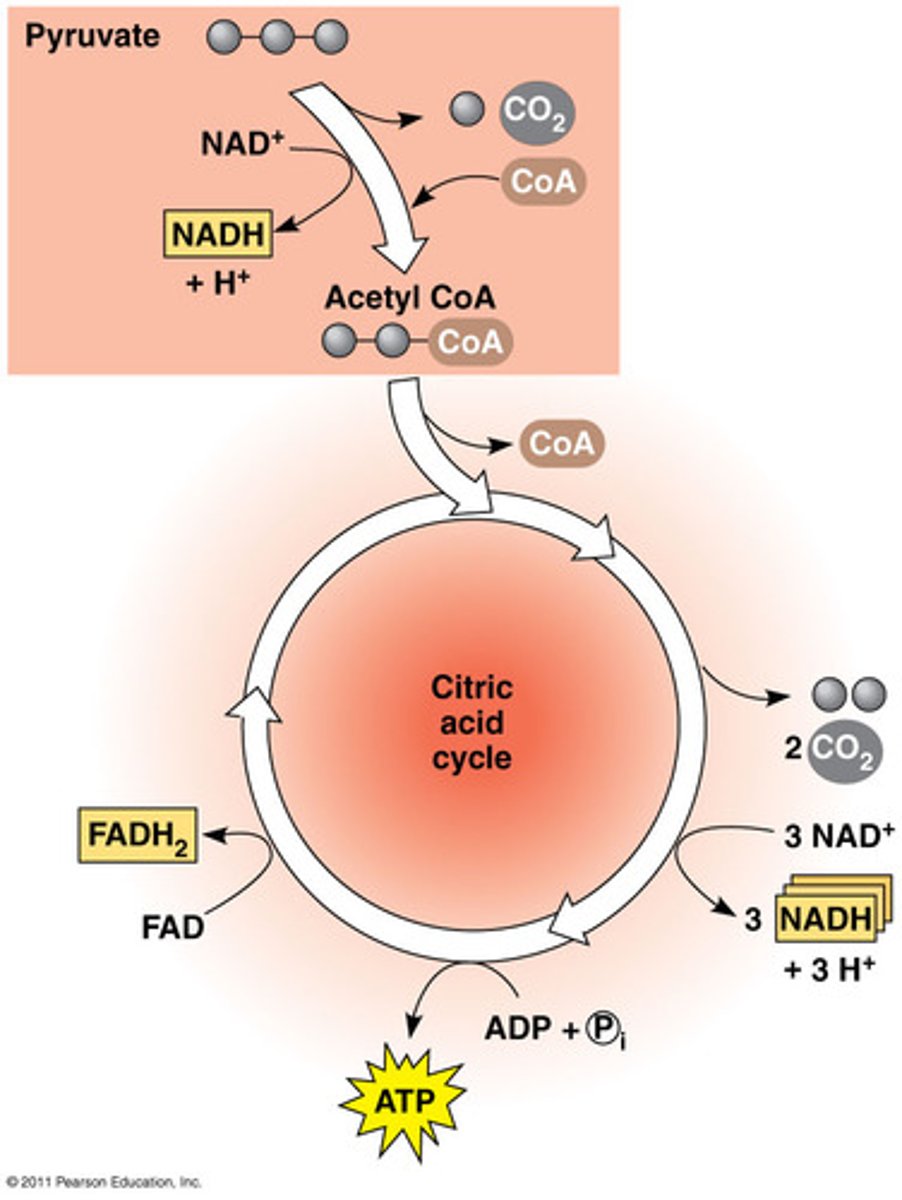
Krebs Cycle
Input: 2 acetyl ➝ citric acid
Output: 2 ATP, 6 NADH, 2 FADH2, 4 CO2 (after 2 turns of the cycle)
Electron Transport Chain
Input: NADH, FADH2, O2 (to accept e-)
Output: 34-38 ATP, H2O
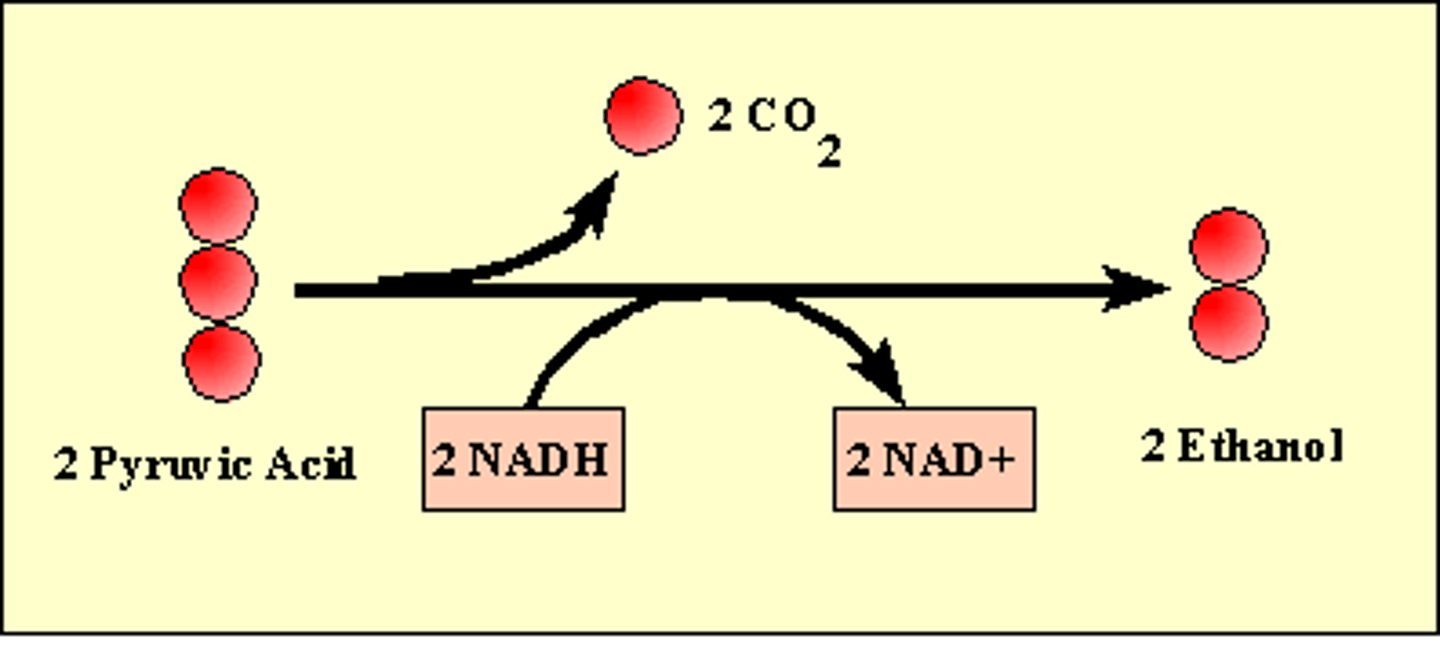
Alcohol Fermentation
Input: glucose, 2 ATP, 2 NADH
Output: 2 NAD+, 2 ethanol, 2 CO2, 4 ATP (net 2)
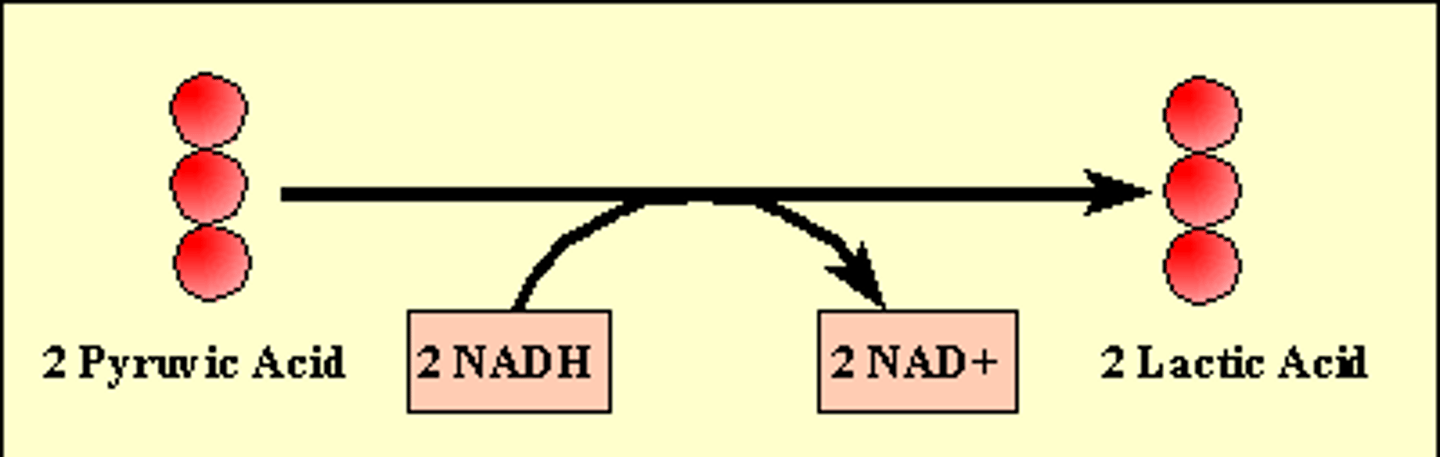
Lactic Acid Fermentation
Input: glucose, 2 ATP, 2 NADH
Output: 2 NAD+, 2 lactate, 4 ATP (net 2)
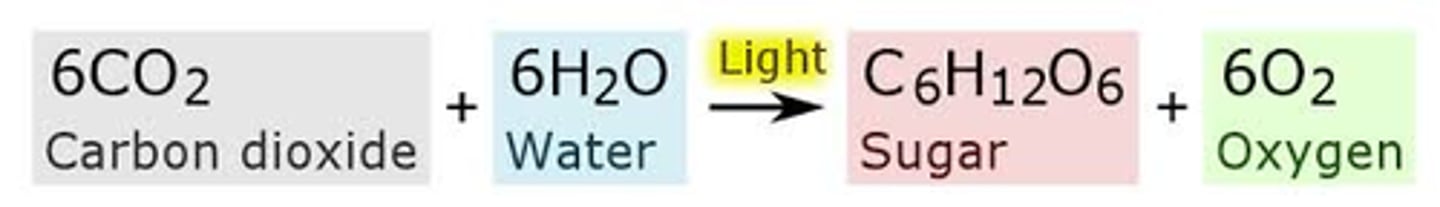
Photosynthetic Equation
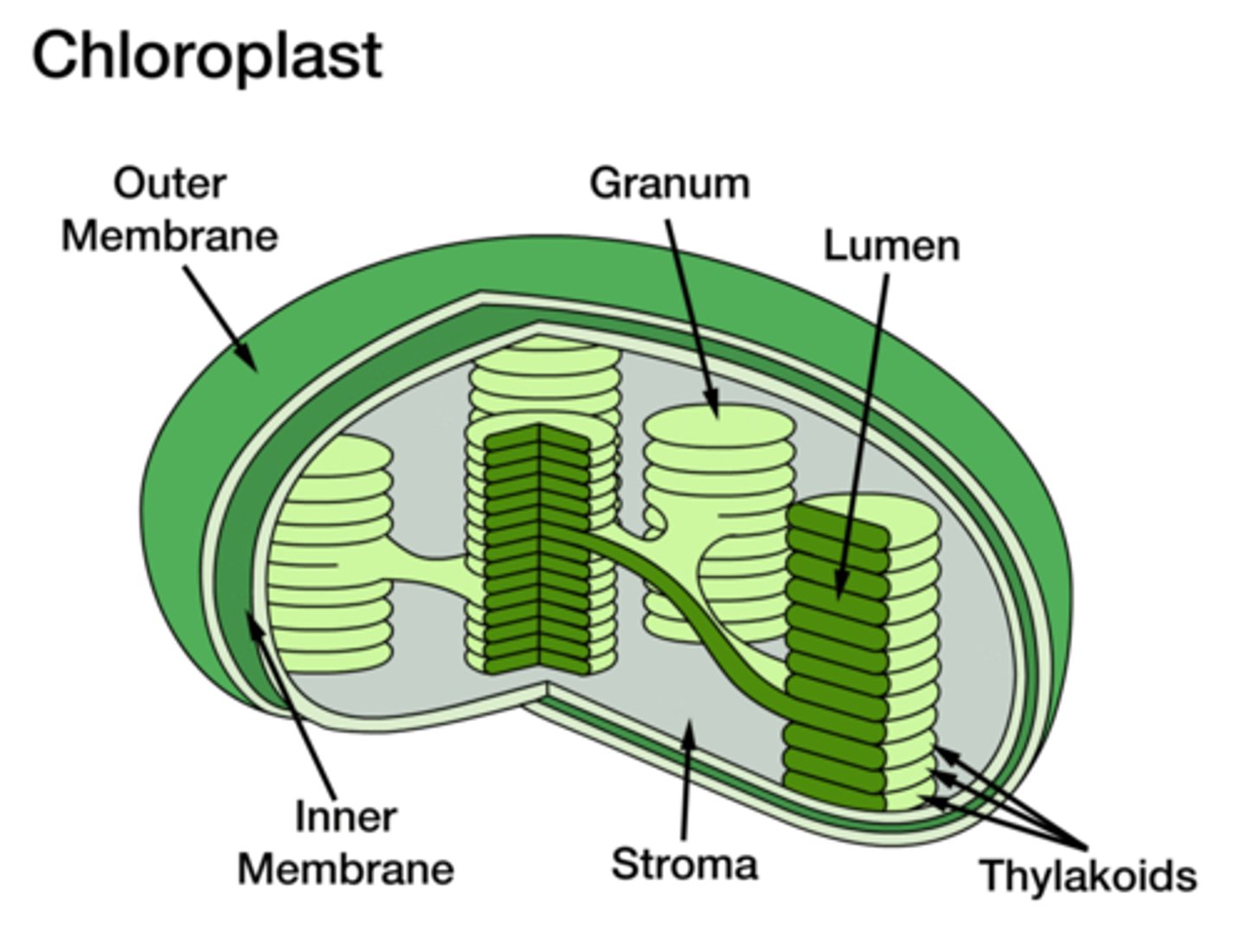
Chloroplast structure
Exciting chlorophyll: chlorophyll in thylakoids absorb light, which excites electrons to produce potential energy
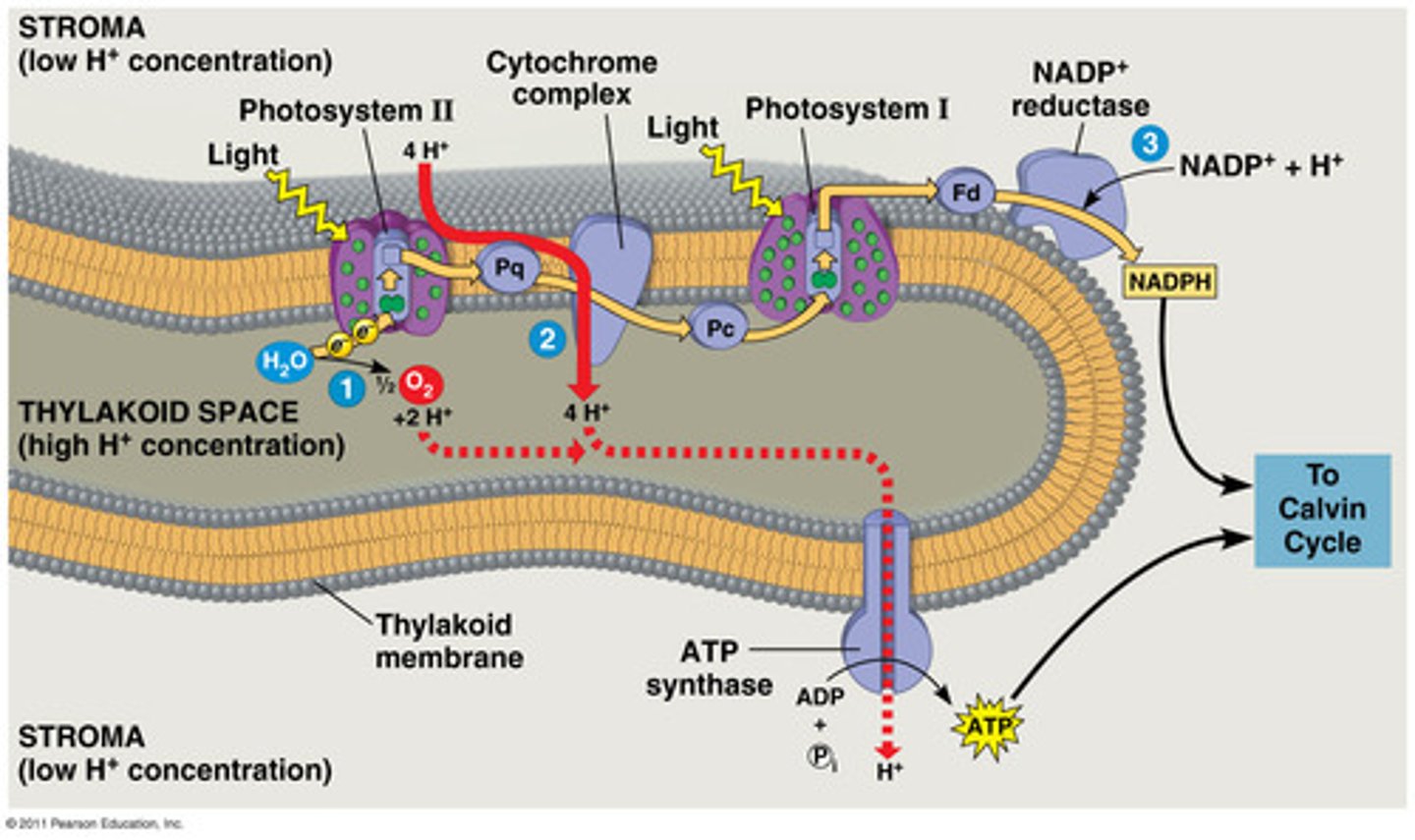
Light Reactions
Input: H2O (2 e-), light energy, NADP+
Output: O2, ATP, NADPH
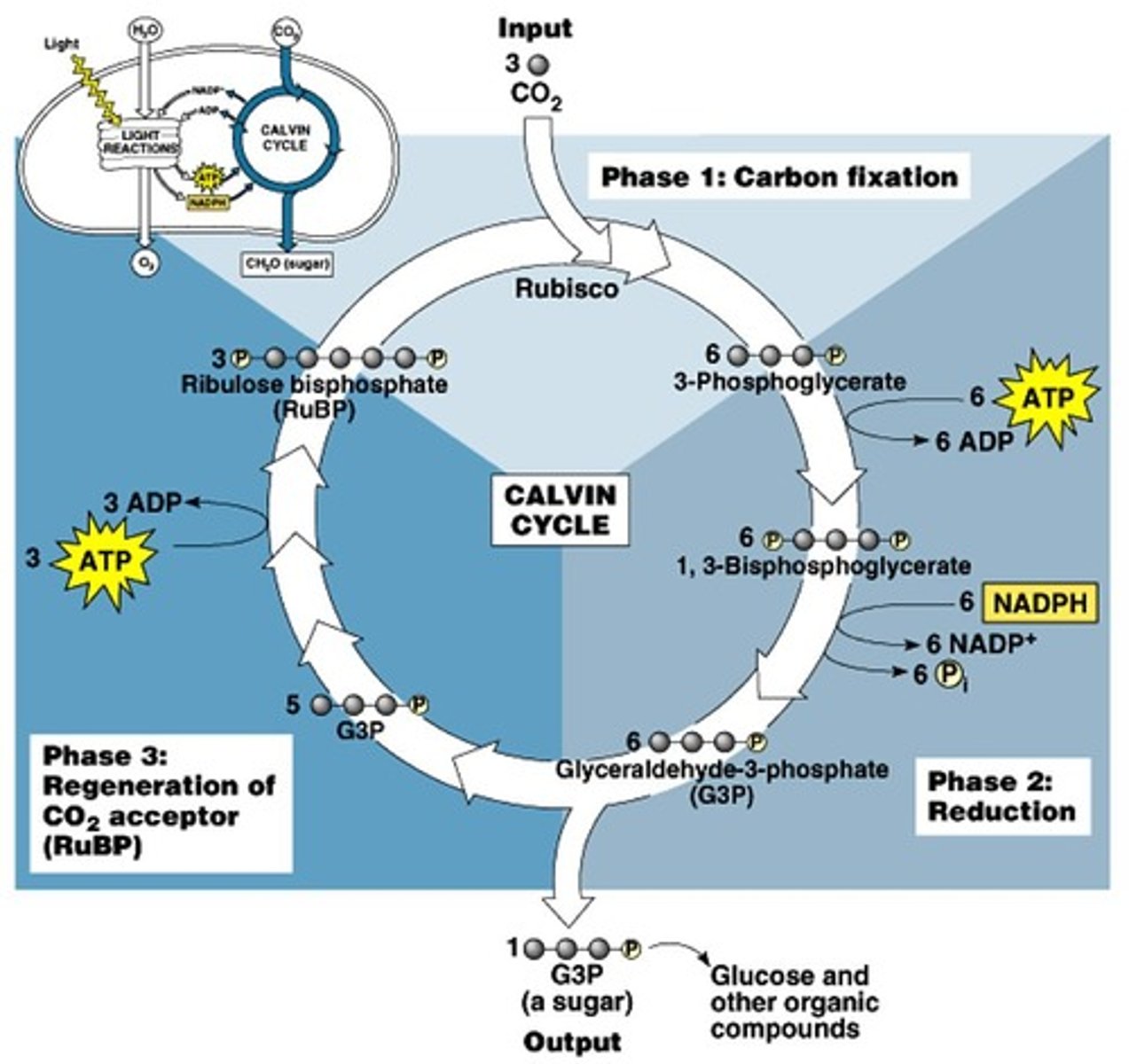
Calvin Cycle
Input: 6 CO2 (fixed to RuBP by Rubisco), ATP, NADPH
Output: 2 G3P = 1 glucose
Watson and Crick
built the first accurate 3D DNA model
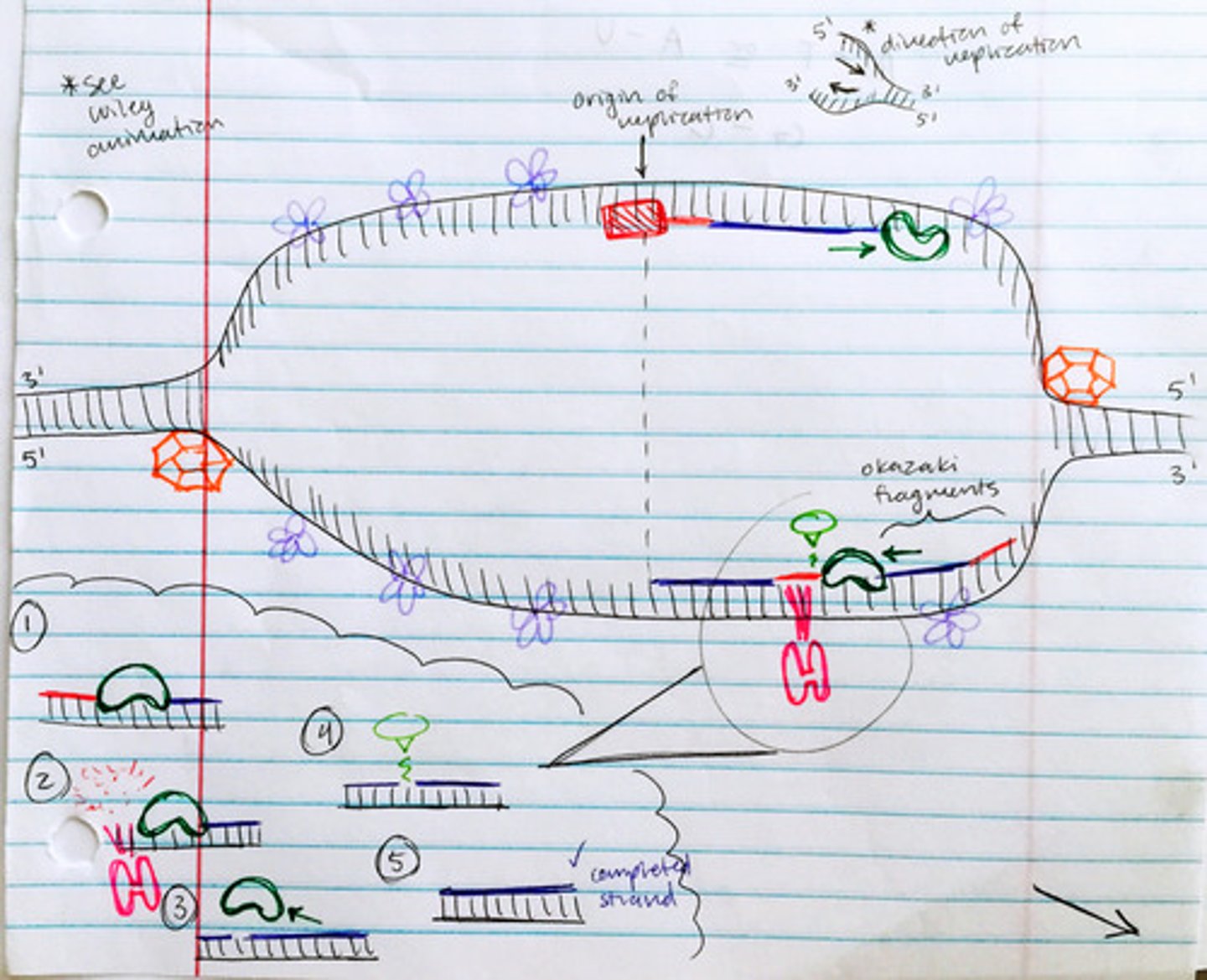
Leading Strand vs. Lagging Strand
works toward replication fork / works away from replication fork; both always move in the 5' ➝ 3' direction
Steps of DNA Replication
1) helicase separates the DNA strands 2) SSB proteins prevent DNA from reanneling 3) primase creates RNA primer 4) DNA polymerase extends DNA strand from the primer 5) DNA polymerase I (RNase H) removes the primers 6) ligase joins the okazaki fragments of the lagging strand
3 types of RNA
1) mRNA messenger 2) tRNA transfer amino acids (20 kinds) 3) rRNA ribosomes
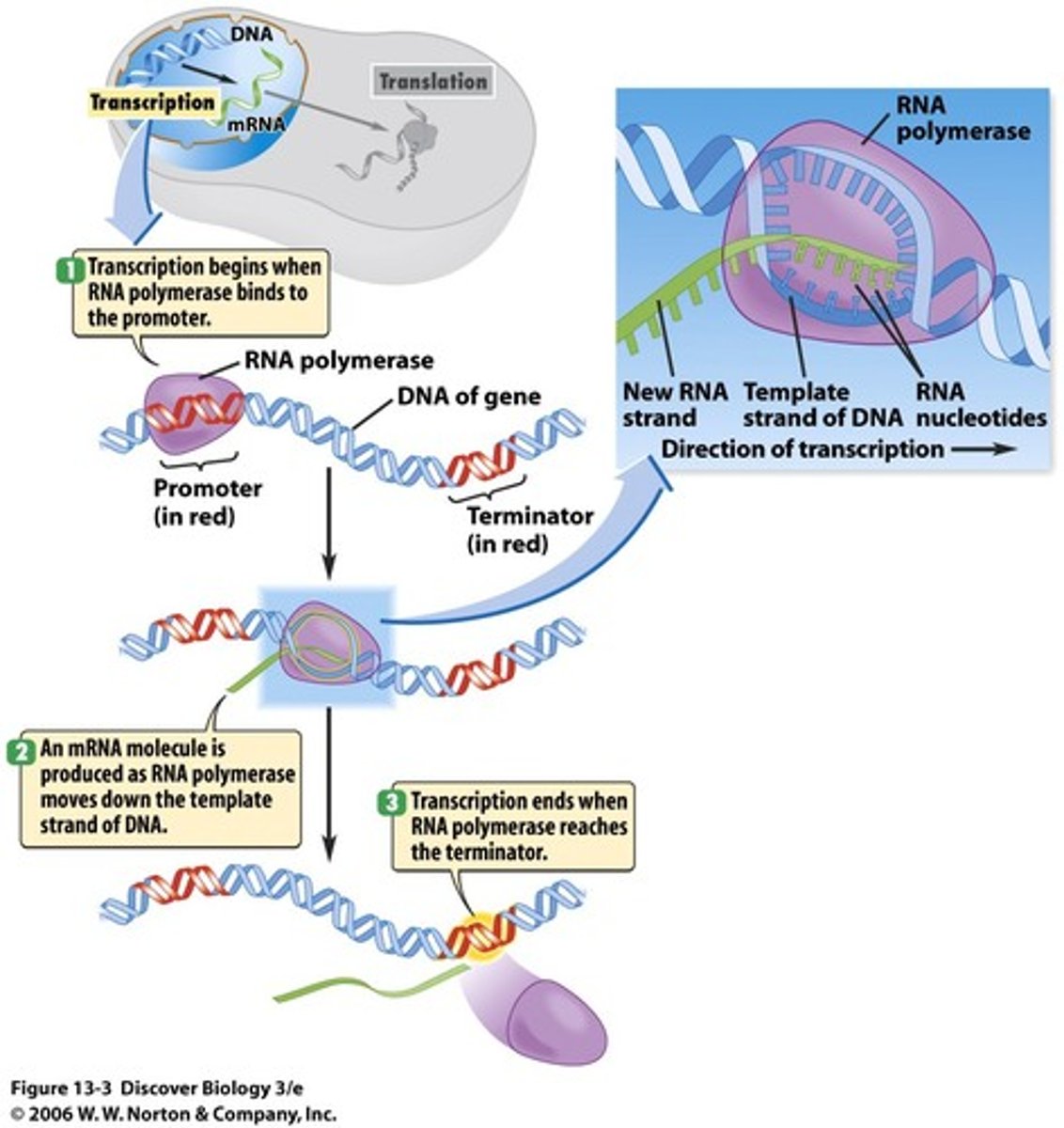
Transcription
1) Initiation: promoter site (TATA) is recognized 2) Elongation: RNA polymerase adds ribonucleotides in the 5' ➝ 3' direction 3) Termination: RNA strand separates, RNA polymerase recognizes termination sequence (AAUAAA)
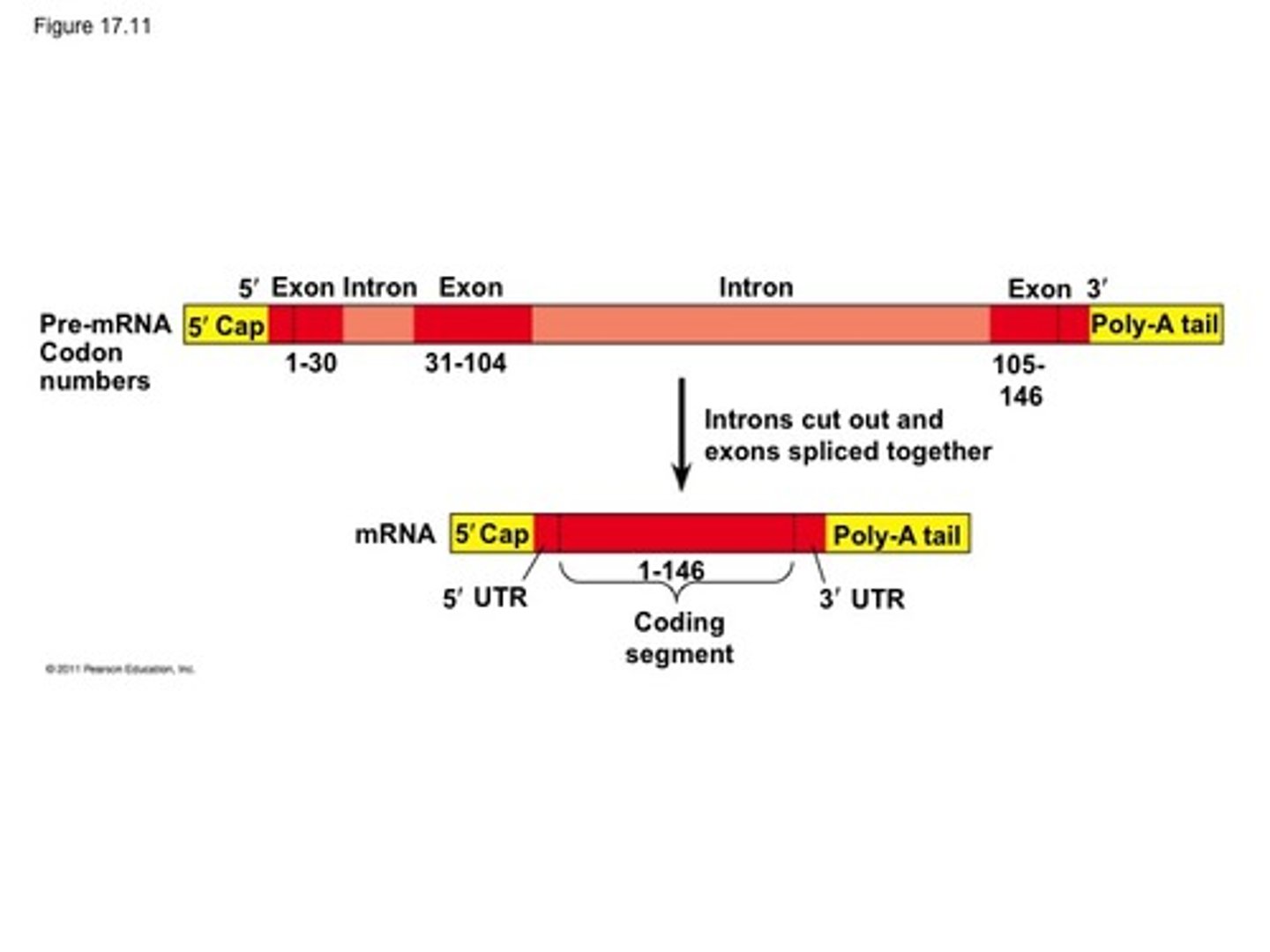
RNA processing/splicing
splicesomes remove introns and put together exons, 5' cap and PolyA tail are added
Codon vs. Anticodon
codon = nucleotide sequence on mRNA
anticodon = nucleotide sequence on tRNA
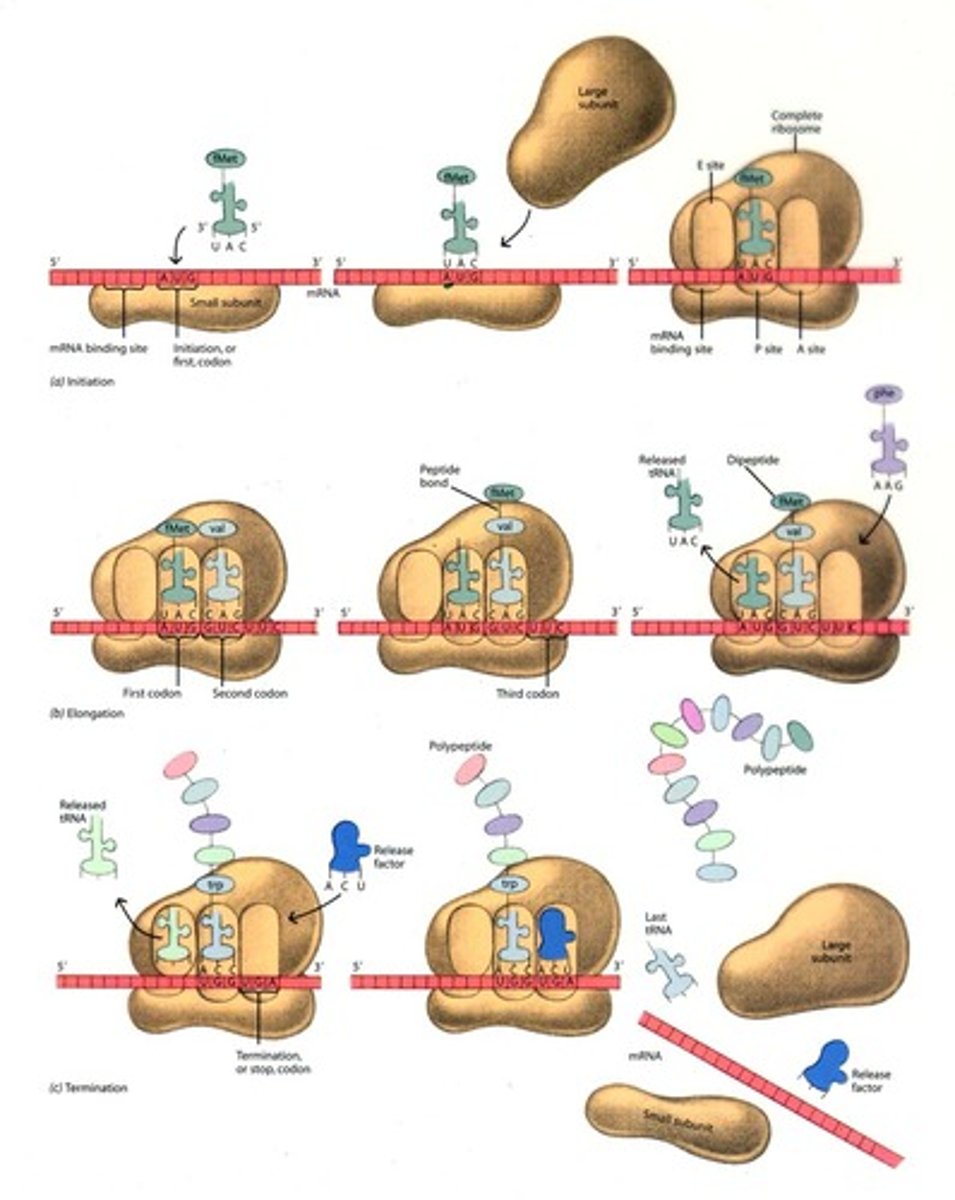
Translation
1) Initiation: 5' cap attaches to ribosome which accepts an initiator tRNA at the P site (*AUG will always be 1st codon) 2) Elongation: codon/anticodon recognition and formation of peptide bond between A site amino acid and P site amino acid chain 3) translocation of the ribosome down the mRNA strand 4) Termination: ribosome will recognize stop codon and release the protein
DNA mutations
base-pair substitution; insertion/deletion; frameshift: 1) missense = different protein 2) nonsense = codes for a stop signal prematurely 3) silent = no harmful change
Prokaryotic cell division
binary fission: splits in 2, exact copies, quick and efficient with few mutations, but reduces amount of genetic variation
Somatic cell vs. Gamete
any body cell except gametes / reproductive cells (sperm, egg)
Interphase
(90% of cell's life) G1: 1st growth, normal metabolic activity (goes into G0 phase if it is not ready for next phase); S: synthesis, DNA replication; G2: 2nd growth, prepares for mitosis
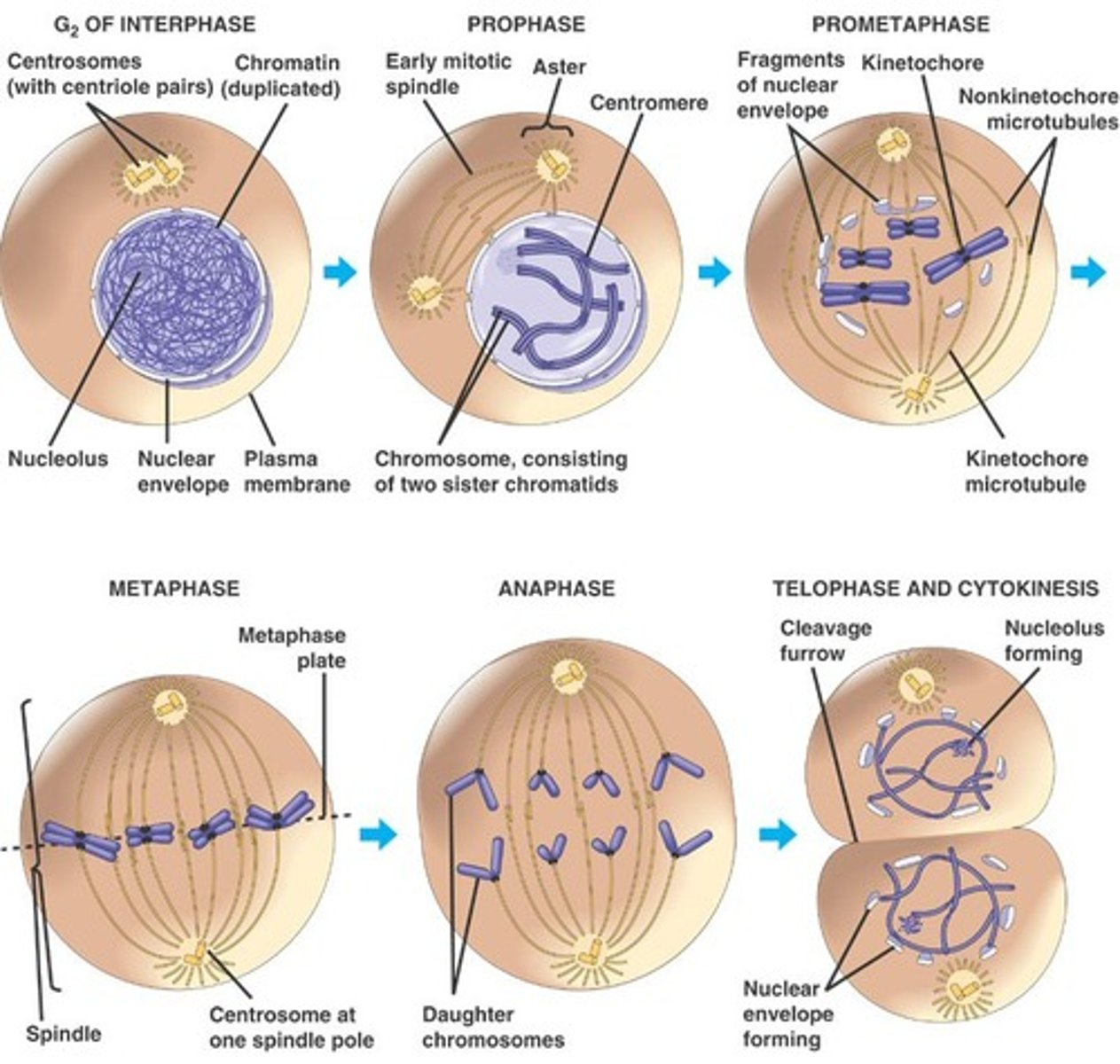
Mitosis
1) Prophase: chromatin condenses into chromosomes, nucleus disappears 2) Metaphase: chromosomes line up at equator, kinetechore microtubules attach 3) Anaphase: sister chromatids move to opposite poles of the cell 4) Telophase and Cytokinesis: daughter cells separate, nucleus reforms, chromosomes decondense
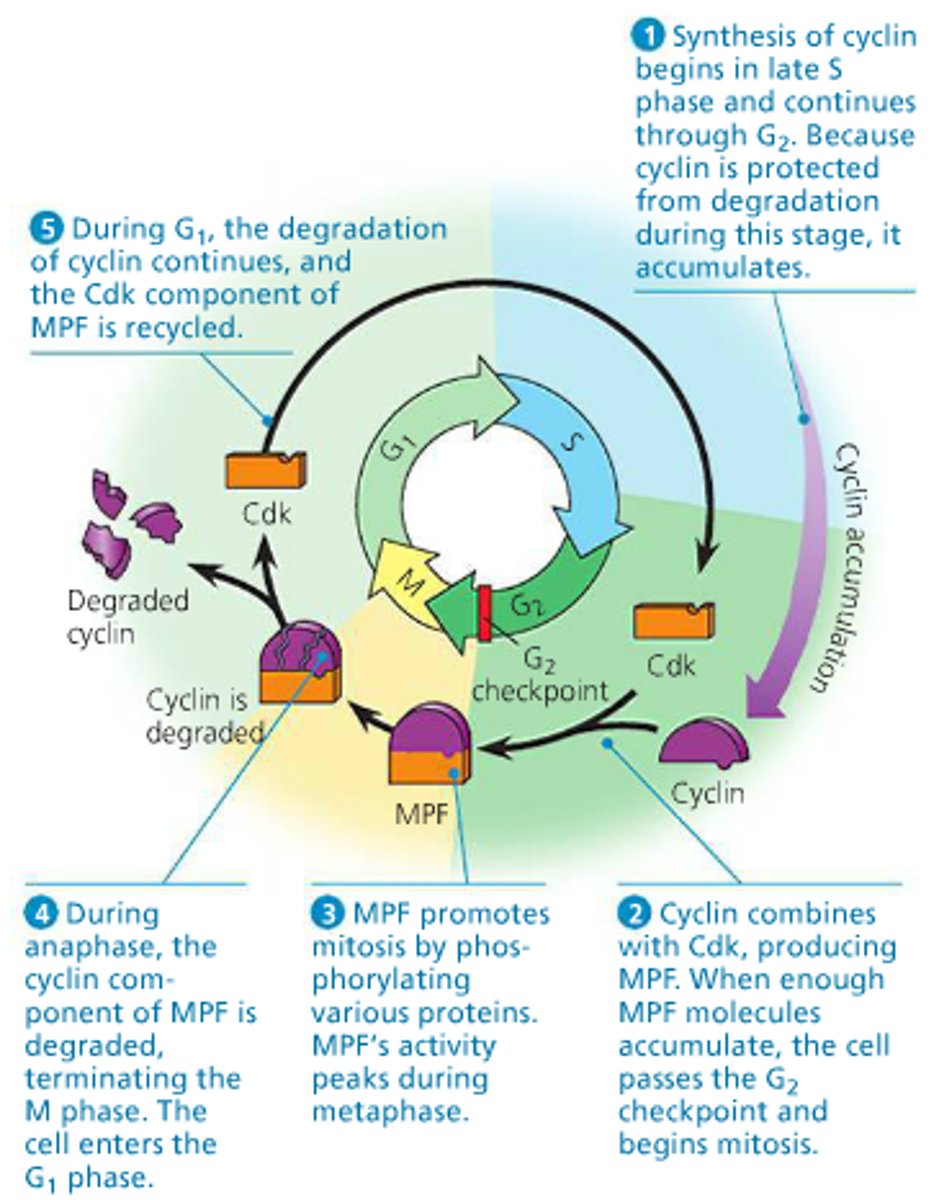
Cyclin-dependent Kinases (Cdks)
a regulatory protein that depends upon the presence of cyclin to complete its function, MPF is a Cdk that triggers a cell's passage into the M phase
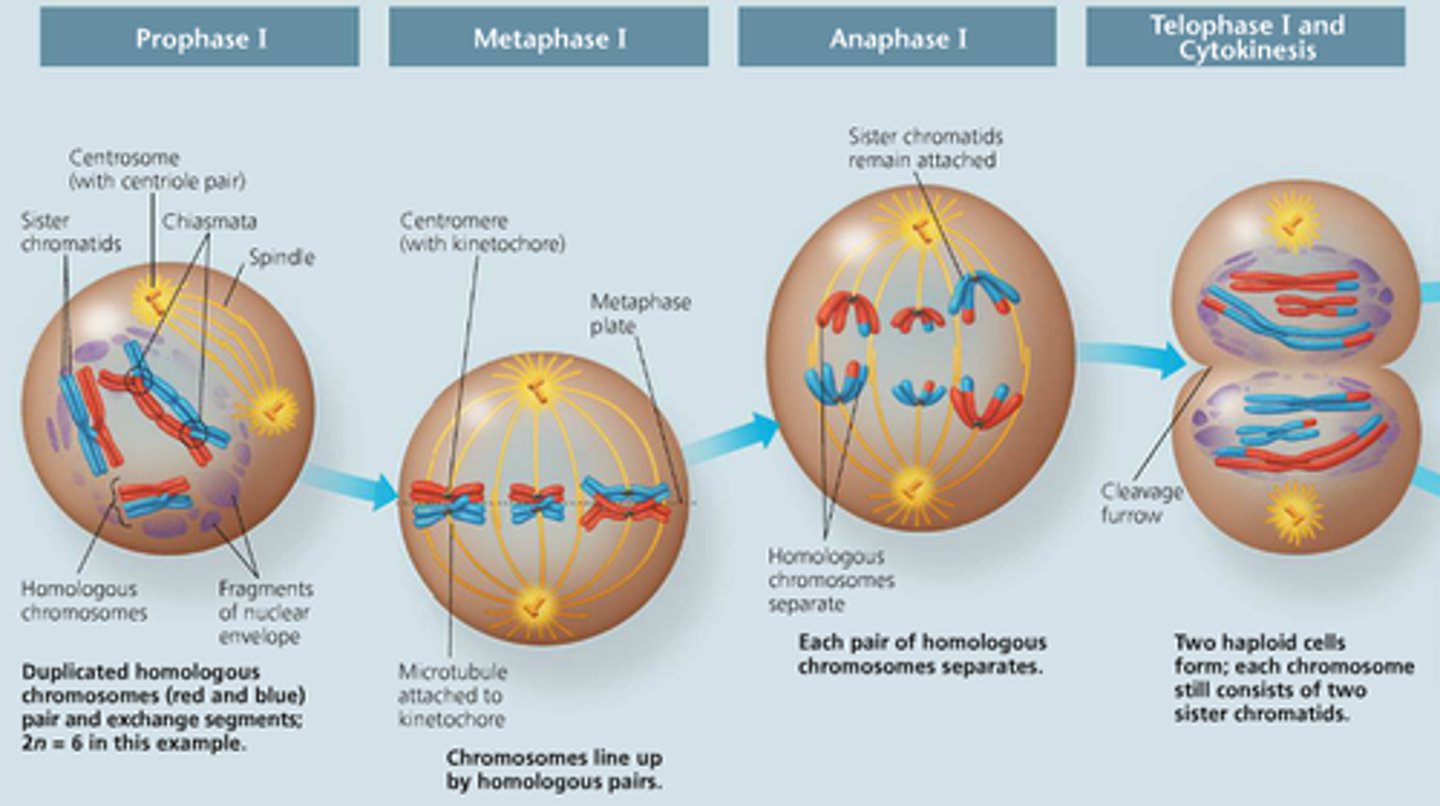
Meiosis I
1) Prophase I: homologous chromosomes pair up and synapsis occurs, crossing over segments of the chromosomes (chiasma) to create more genetic variation 2) Metaphase I: homologous chromosomes line up at the equator 3) Anaphase: homologous chromosomes move to opposite poles of the cell. 4) Telophase I...
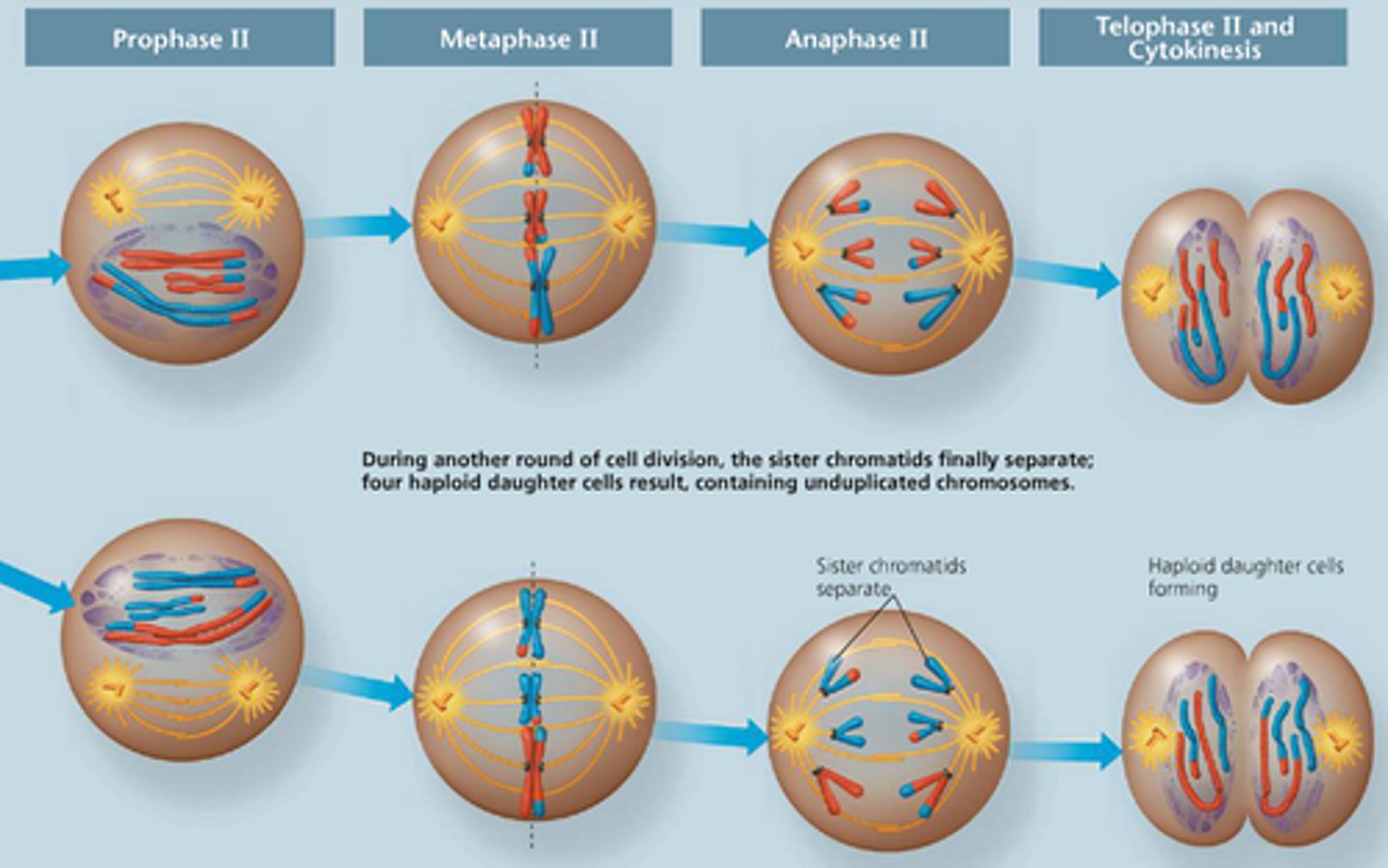
Meiosis II
Prophase II - Telophase II act exactly like mitosis except that the resultant number of daughter cells is 4 instead of 2, each with their own unique combination of genetic information
4 mechanisms that contribute to genetic variation
1) Mutation 2) Independent Assortment: homologous chromosomes align randomly on one side of the equator or another 3) Crossing Over 4) Random Fertilization: a zygote can be any combination of a sperm and egg (64 trillion different combinations in humans)
Testcross
breed a homozygous recessive individual with an individual with a dominant phenotype but an unknown genotype to determine whether or not the individual is homozygous or heterozygous
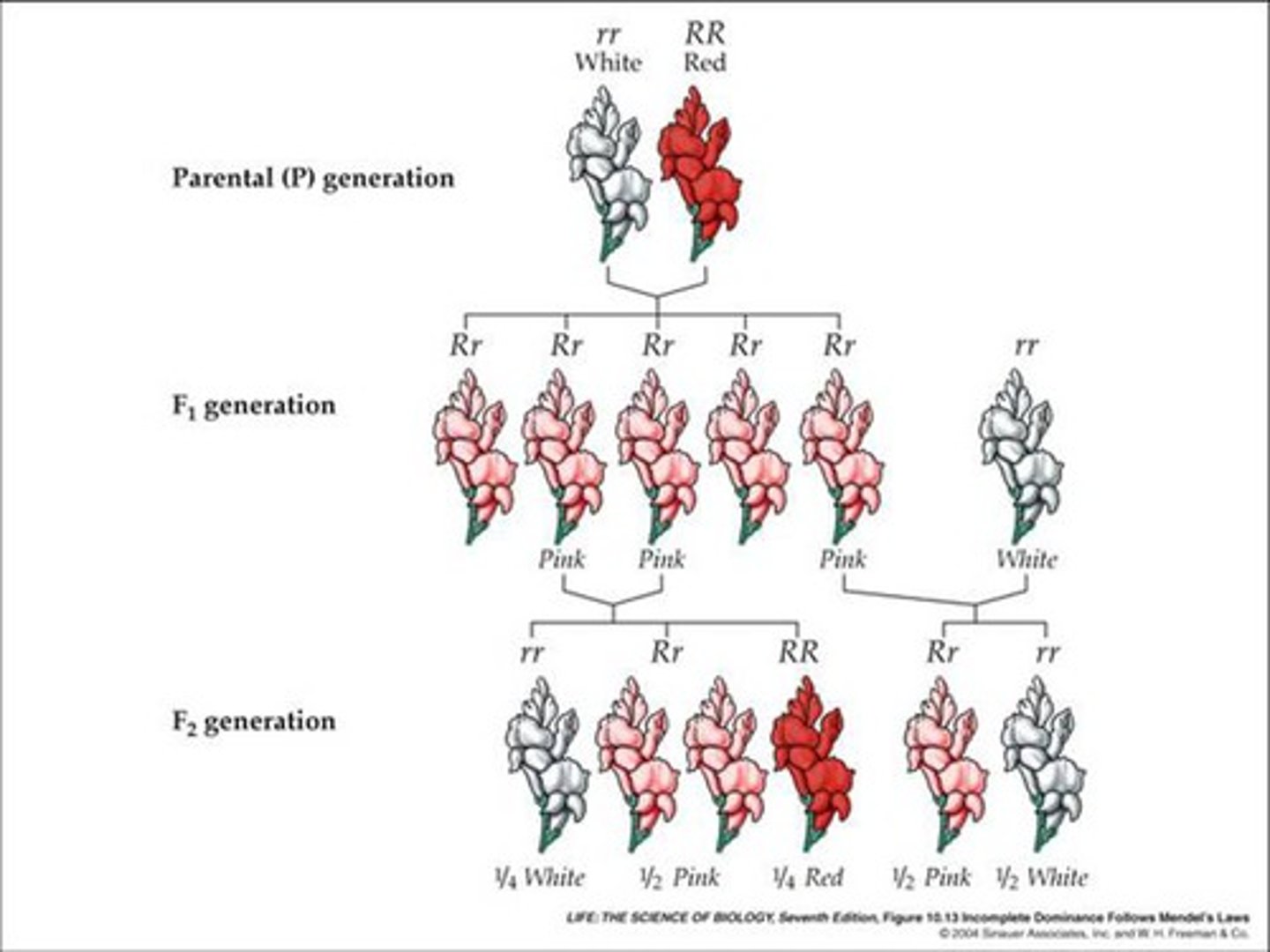
Incomplete Dominance
heterozygous offspring have an intermediate phenotype of the parents, 1:2:1 ratio (ex. pink flower from red and white flowers)
Codominance
both alleles manifest themselves separately in an organism's phenotype (ex. roan cattle)

Multiple alleles
a trait controlled by two or more alleles (ex. blood type, eye color)

Blood Types
A: A antigen, B antibody
B: B antigen, A antibody
AB: A and B antigen, no antibodies (universal recipient)
O: no antigens, A and B antibodies (universal donor)
Linked genes phenotypic ratio
two large numbers (wild and mutant) and two much smaller numbers (recombinant phenotypes)
Genetic Map (Linkage/Cytological Map)
ordered list of the genetic loci along a particular chromosome, recombinant frequencies can be used to construct it (smaller the percentage = closer together)
X Inactivation
in females during embryonic development, one of the two X chromosomes in a cell becomes inactive (Barr body) (ex. calico cats)
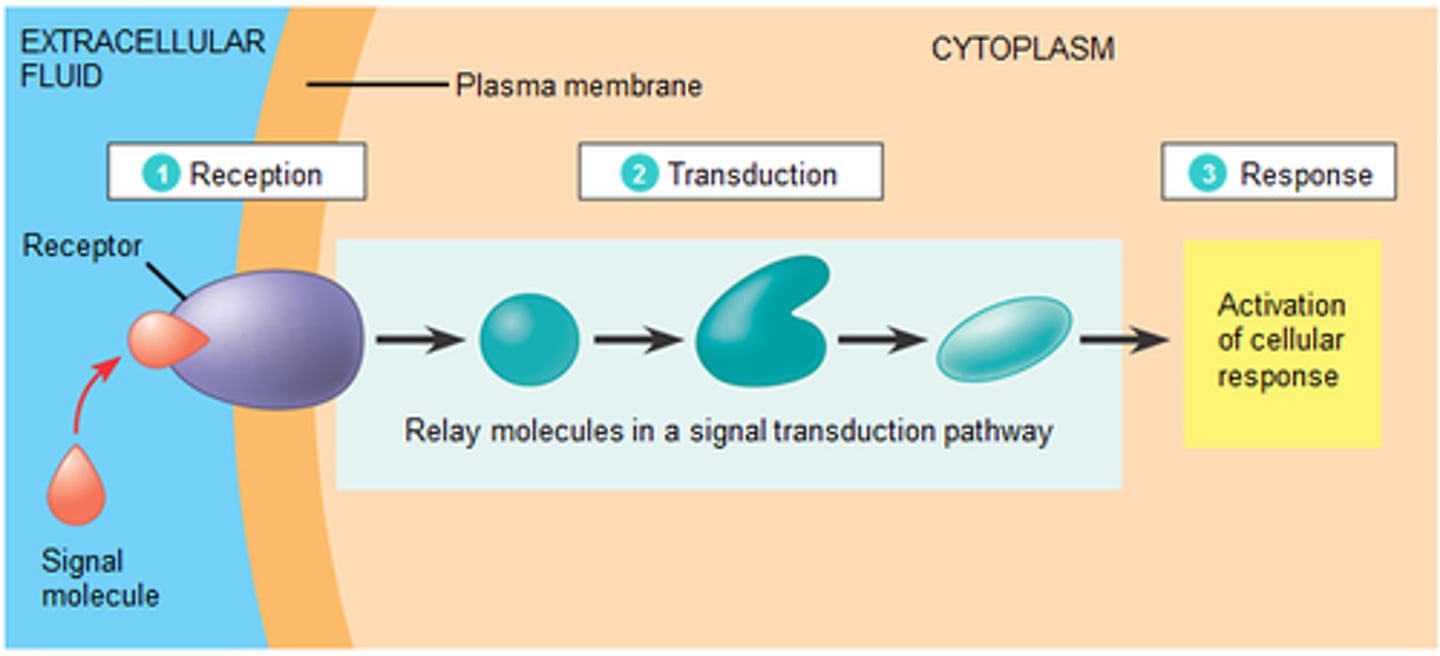
3 stages in cell communication
1) Reception: cell detects a signal via connection of a ligand to a receptor protein 2) Transduction: the receptor protein converts the signal to a form that can cause a chemical response 3) Response: transduced signal triggers a specific cellular response
Types of cell signaling (3)
synaptic, paracrine, hormonal
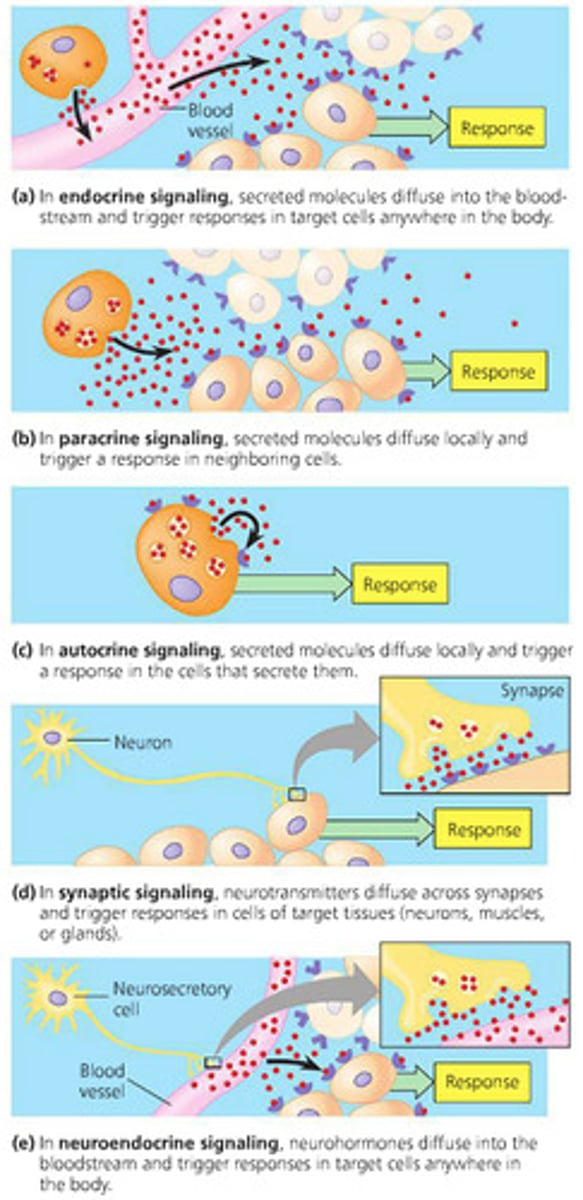
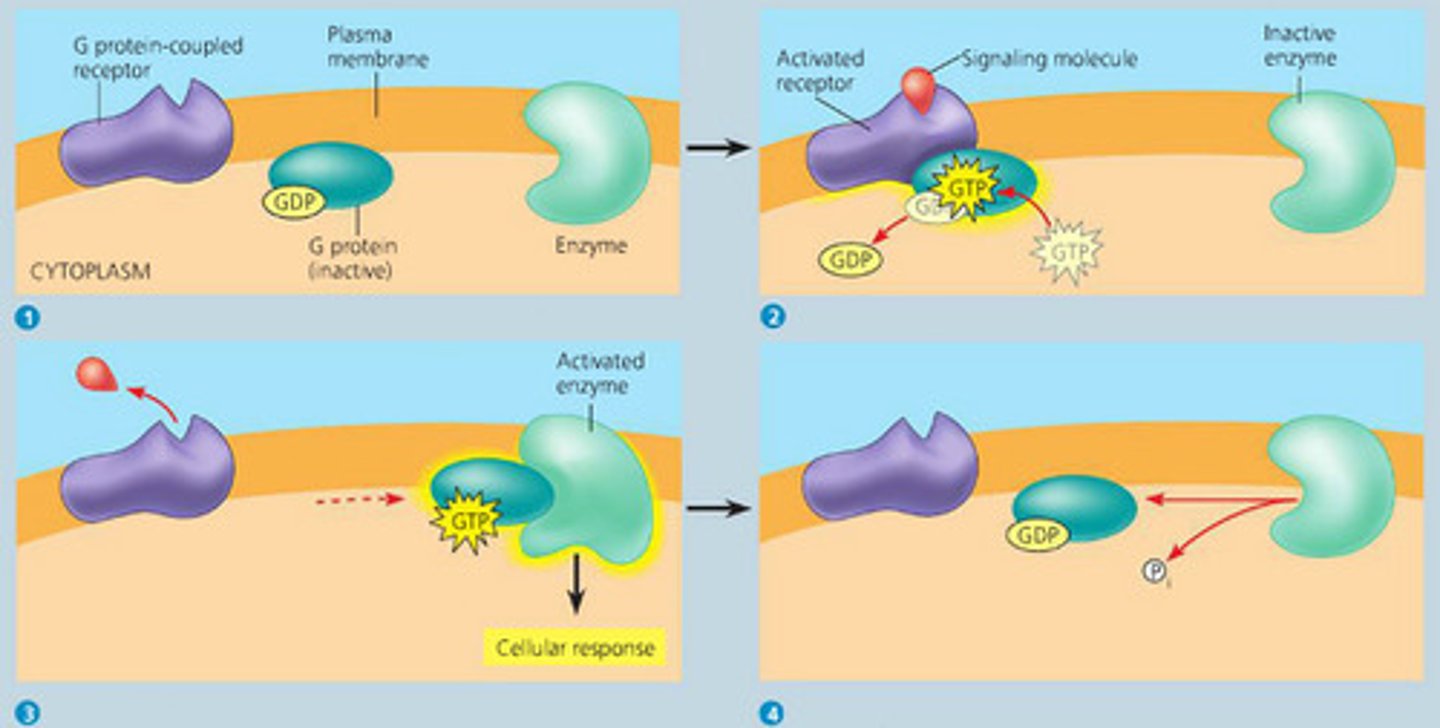
Examples of cell signaling
G-protein coupled receptor, ligand-gated ion channels, steroid hormones (dissolved across plasma membrane, intracellular receptor)
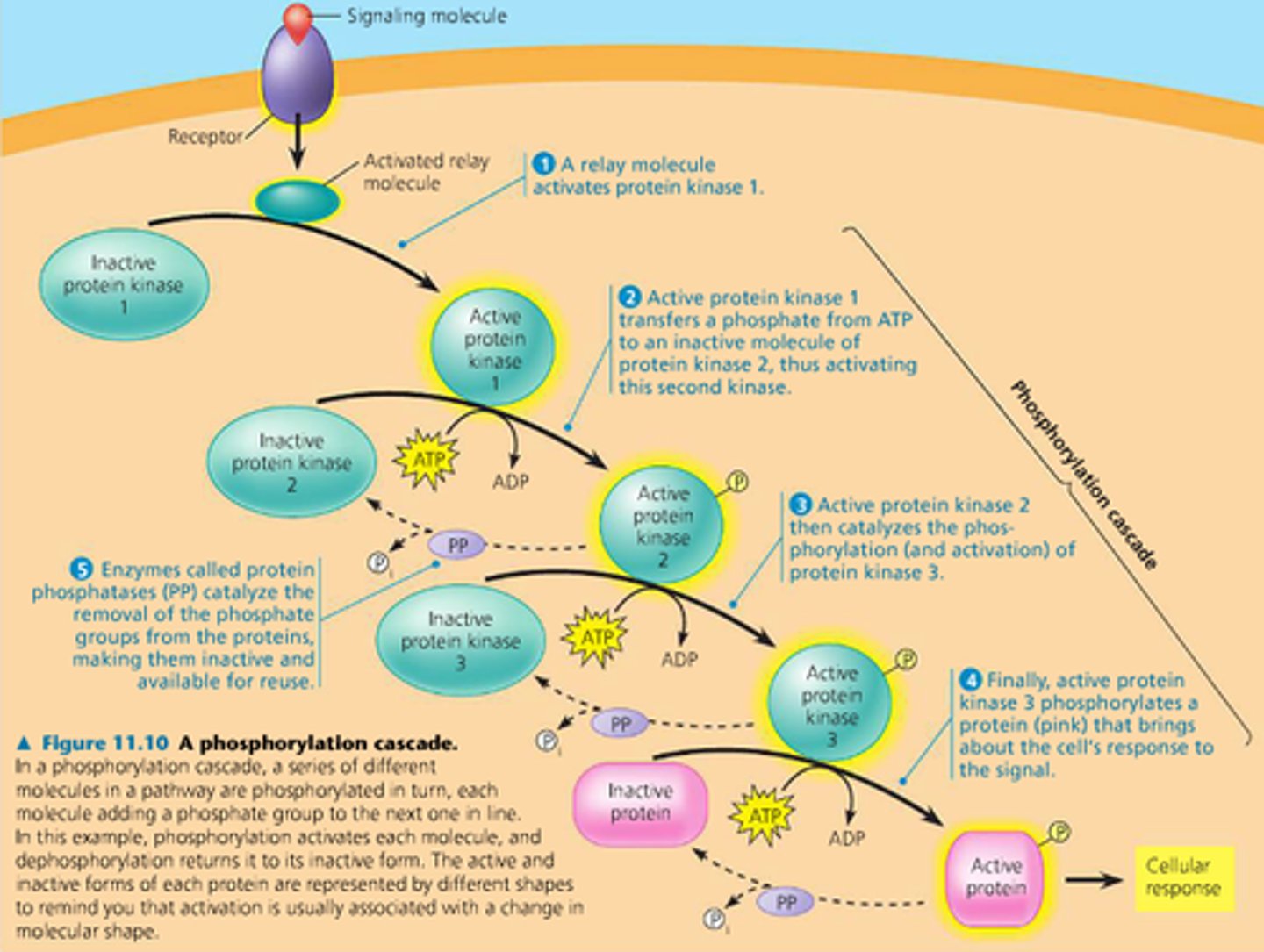
Second Messengers and Phosphorylation cascade
second messengers and kinases spread throughout a cell that help amplify a cellular signal by a series of phosphorylation reactions (addition of phosphate)
Evidence for Evolution
1) Biogeography 2) Fossil Record 3) Comparative Anatomy 4) Comparative Embryology 5) Molecular Biology
4 conditions for Hardy-Weinberg Equilibrium (not evolving)
1) very large population 2) isolation from other populations 3) no mutations 4) no natural selection
Microevolution vs. Macroevolution
change in the gene pool of a population over several generations / large scale changes in a population that leads to the evolution of a new species
4 causes of Microevolution
1) genetic drift 2) gene flow 4) natural selection
Genetic Drift
random change in gene frequency of a small breeding population: 1) Founder Effect = small population of organisms colonizes a new area, 2) Bottleneck Effect = sudden decrease in population size due to disaster
Gene Flow
loss/addition of alleles from a population due to imigration/emigration
Nonrandom Mating
selection of mates for specific phenotypes: 1) Assortative Mating = when individuals select partners with simple phenotypic characters, 2) Inbreeding = more recessive traits likely to come together
3 Modes of Natural Selection
1) Stabilizing: favors intermediate, 2) Directional: favors one extreme phenotype, 3) Diversifying: favors both extremes
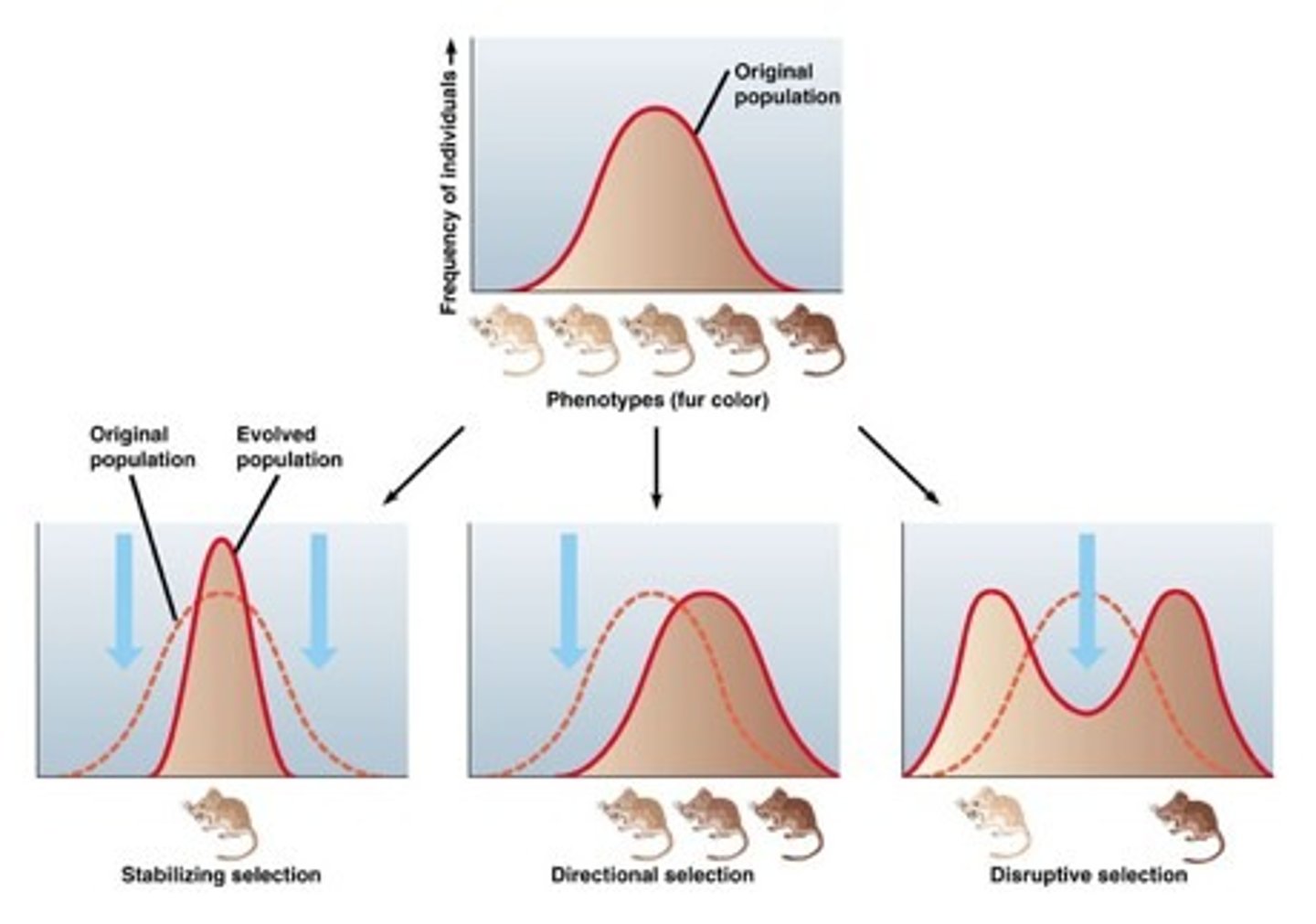
Heterozygote Advantage
heterozygotes for a trait are more likely to survive (ex. carriers of sickle cell anemia are immune to malaria)
Biological Species Concept
population whose members can create viable, fertile offspring (Problems: doesn't apply to extinct animals or asexually reproducing organisms)
Prezygotic Reproductive Barriers
1) Habitat Isolation 2) Behavioral Isolation (differing behaviors for attracting mates) 3) Temporal Isolation (mate at different times) 4) Mechanical Isolation 5) Gametic Isolation (unable to fertilize egg)
Postzygotic Reproductive Barriers
1) Reduced Hybrid Viability (disruption in embryonic stage) 2) Reduced Hybrid Fertility 3) Hybrid Breakdown (F1 is fertile, F2 is sterile or weak)
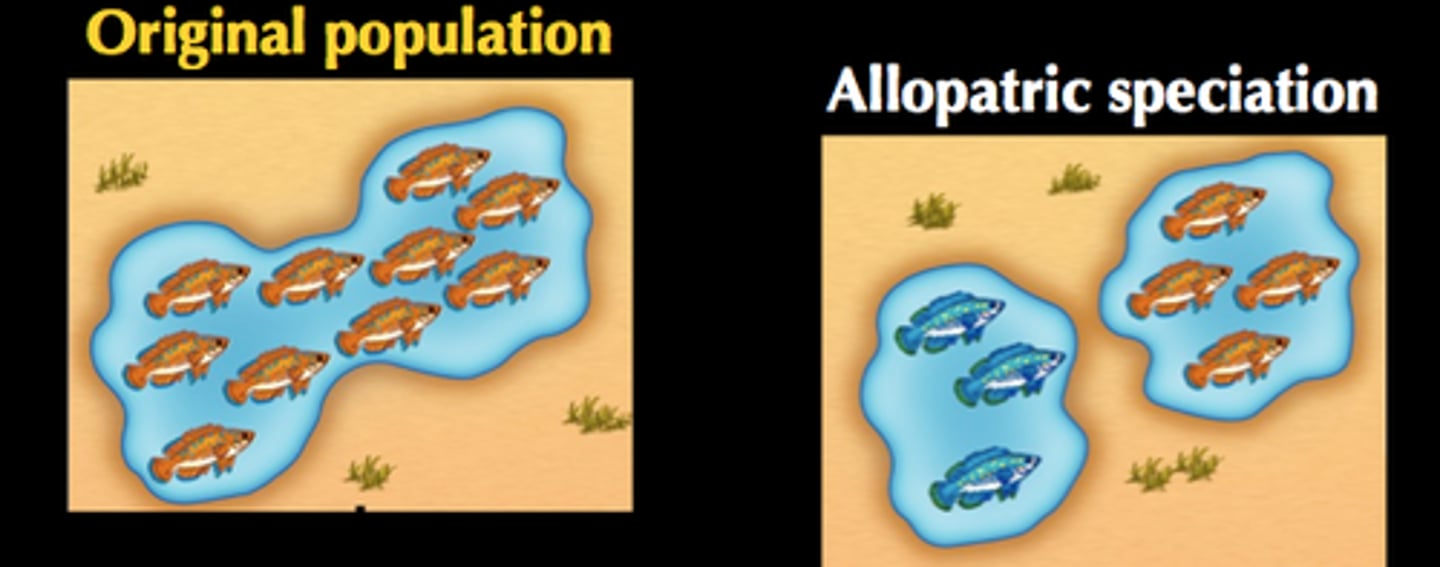
Allopatric Speciation
when populations become geographically isolated from the rest of the species and has the potential to develop a new species (ex. Adaptive Radiation: many diversely adapted species from common ancestor, Darwin's finches)