L4: Eukaryotic chromosome replication II
1/40
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
41 Terms
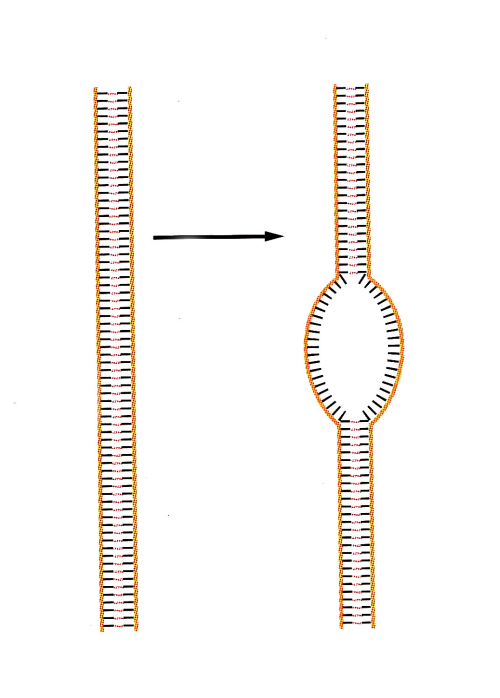
What is the first enzymatic step of DNA replication
localised separation
or unwinding of the two DNA strands at the replication origin
catalsed by DNA helicases
Unwound DNA is stabilised by single-stranded binding proteins
DNA polymerases and additional proteins are recruited thhat built up active DNA replication forks
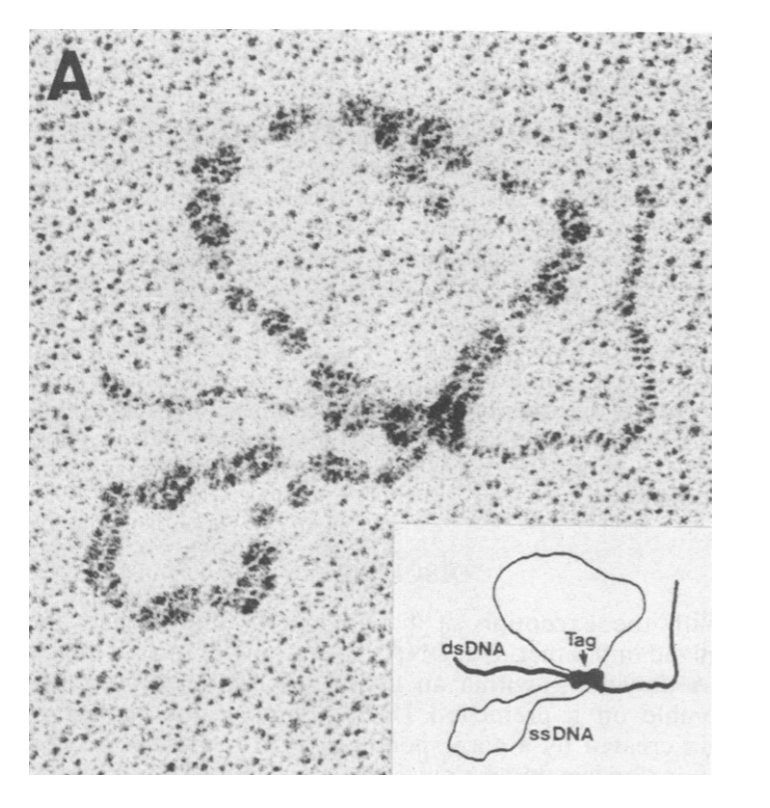
This is seen in SV40 origin unwinding
with large T antigen
The replicative DNA helicase: the eukaryotic helicase is…
a core complex of six MCM proteins
with many other associated proteins:
Cdc45
GINS complex
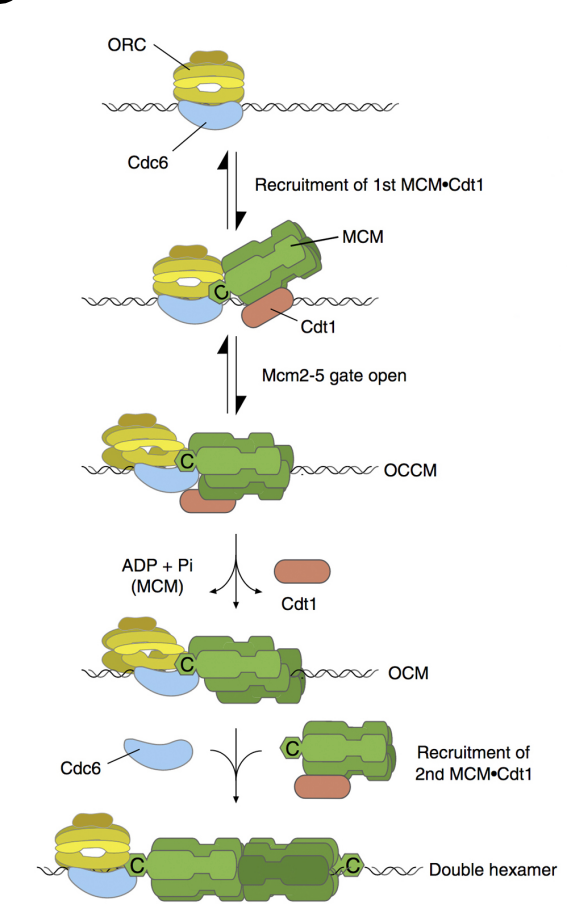
Loading the MCM complex (the helicase)
MCM double hexamer complex is loaded at replication origins in an ATP-dependent manner
by origin recognition complex ORC
Involving Cdc6 and cdt1 proteins
(ORC binds to Cdc6→ 6 subunits hexamer complex)
Loads Mcm on DNA
The complex can now move along DNA→ similar to what homohexmaer in T antigen does
Removes cdt1 (accessory protein)
recruits another copy of the hexamer (again using a cdt1 to help it load)
The double hexamers can move away from eachother (but have to be activated first)
Activation of the MCM helicase in S phase
Loaded but inactive MCM double hexamer complex is converted into an active form
With protein kinases CDK and DDK
with association with several other proteins:
cdc45 and GINS DDK pol epsion (pol subunit)
Complex: CMG complex
(Cdc45, MCM, GINS)
Mcm10 added
origin melting,
MCM ring opening,
Double hexamer separation
Helicase activation
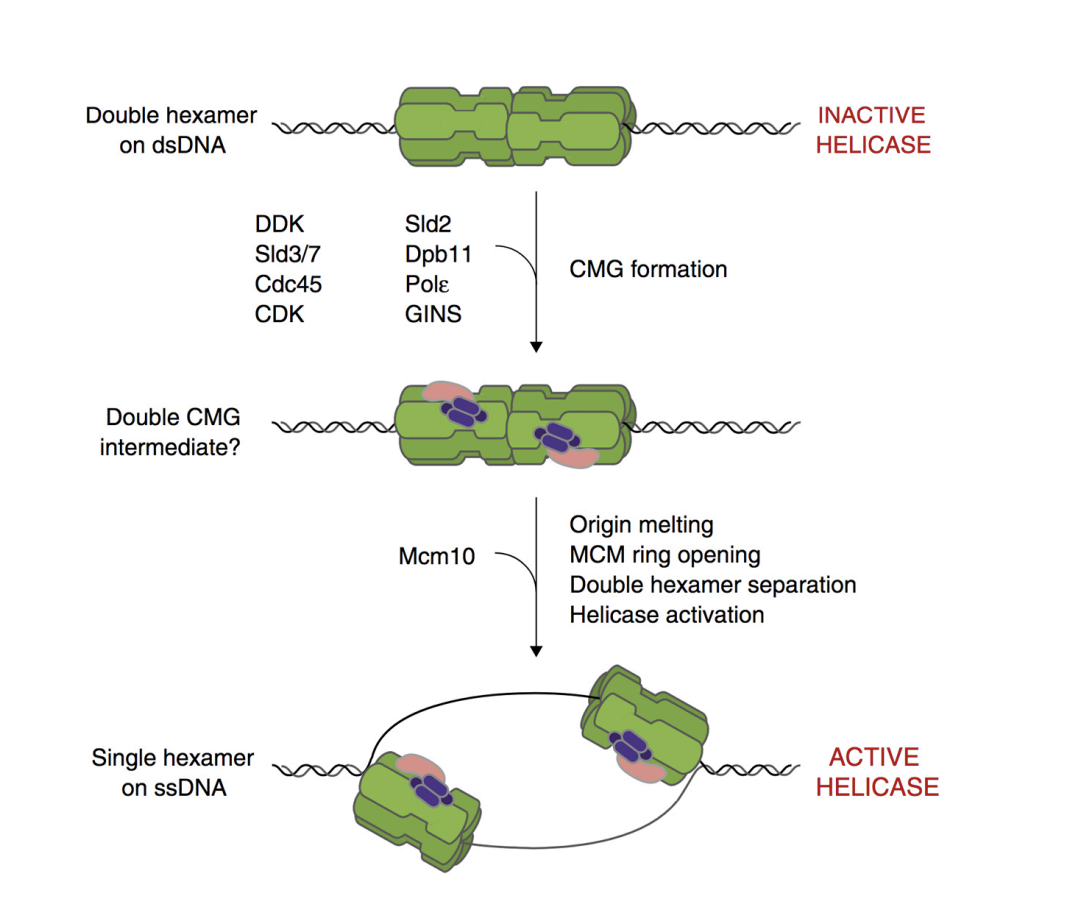
What does DNA helicase activation lead to
local unwinding
separation of the double hexamer complexes
ATP used to migrate and push
Where do each of the two CMG helicase complexes travel
travel with one of the two emerging replication forks away from the initiation site
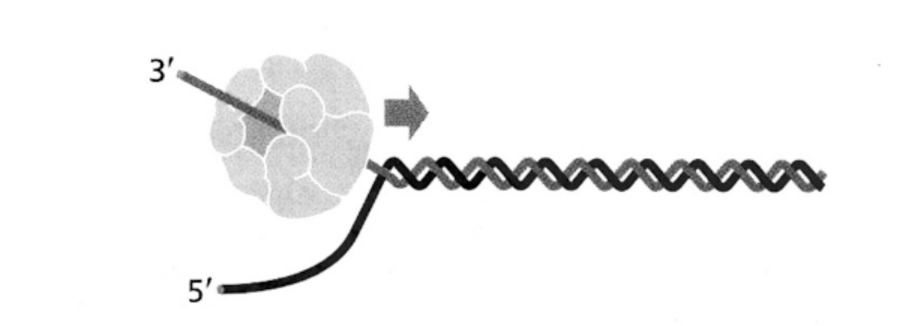
After unwinding, the active CMG DNA helicase…
translocates on the DNA leading strand
in 3’ to 5’ direction
Dependent on ATP hydrolysis
THEREFORE→ displacing the complementary DNA strand
What does this cause?
Around activated helicases, functional DNA replication fork complexes are assempled
Involving the recruitment of the DNA polymerases and replication factors
Conservation
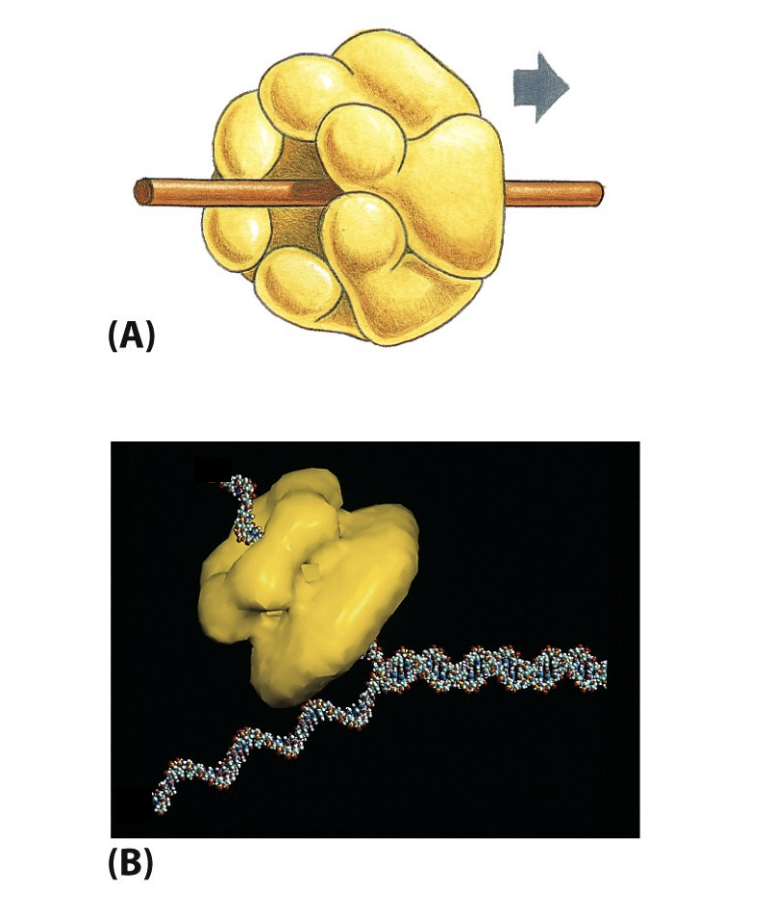
Strucutre of eukaroytic DNA helicase:
E1 protein of papillomavirus and ssDNA
similar strucutres have been obtained for the cellular MCM2-7 hexamer complexes
NOTE: no origin consensus sequences in vertebreates (unlike in yeast) BUT ORC cdc6, cdt1 and MCMs are strucutrually and functionally concerved from yeasts to vertebrates
How we know about protein funtions
mutant genetics
biochem→ reductionist→ until pure protins and so test function
Comparing SV40 viral genome to eukaroytic cellular genomes
SV40 viral genome
one protein for both binding and helicase activity
Eukaryotic cellular genomes
two protein complexes:
one for origin binding and helicase loading (ORC etc)
one for helicase acitivity (MCM/CMG etc)
Aims of DNA replication
Types of DNA polymerases→ 6 major DNA polymerases (pol) in eukaryotes
polyermisation in 5’ to 3’ direction by DNA polymerases
→ because the chemistry of DNA is uni-directional
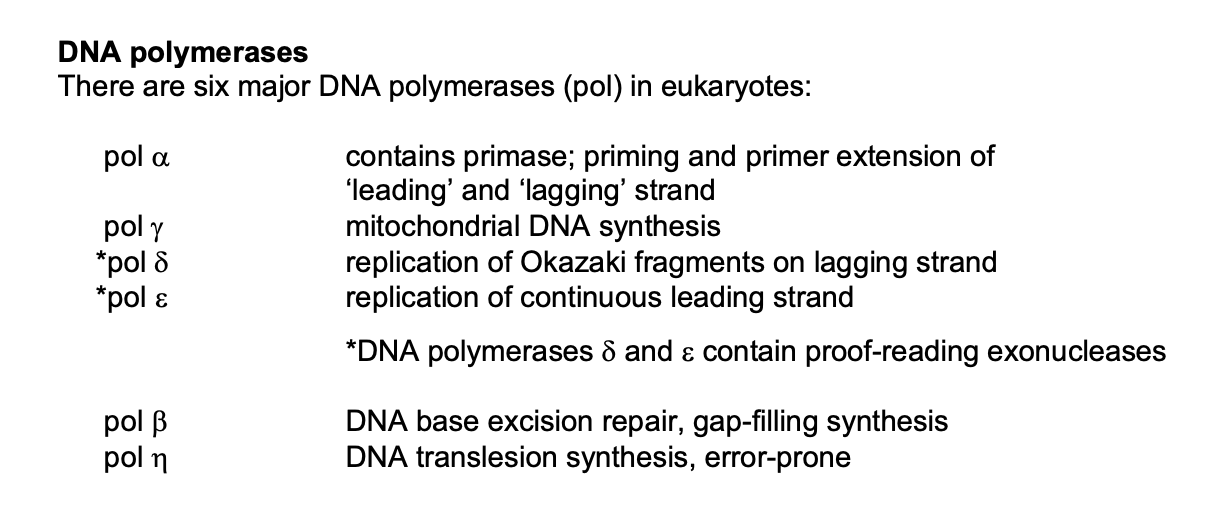
Three main types of DNA polymerases
alpha
contains purine
needed for priming (RNA synthesis)
limited primer extension (DNA synthesis)
→ starts off the replication
Delta
Highly processive replication
lagging strand synthesis (Okazaki fragments)
→ helps complete the replication coz it can do longer stretches
Epsilon
highly processive replication
leading strand synthesis
→ helps complete the replication in the LEADING
DNA polyermases delta and epsilon contain:
proof-reading exonuclease activity
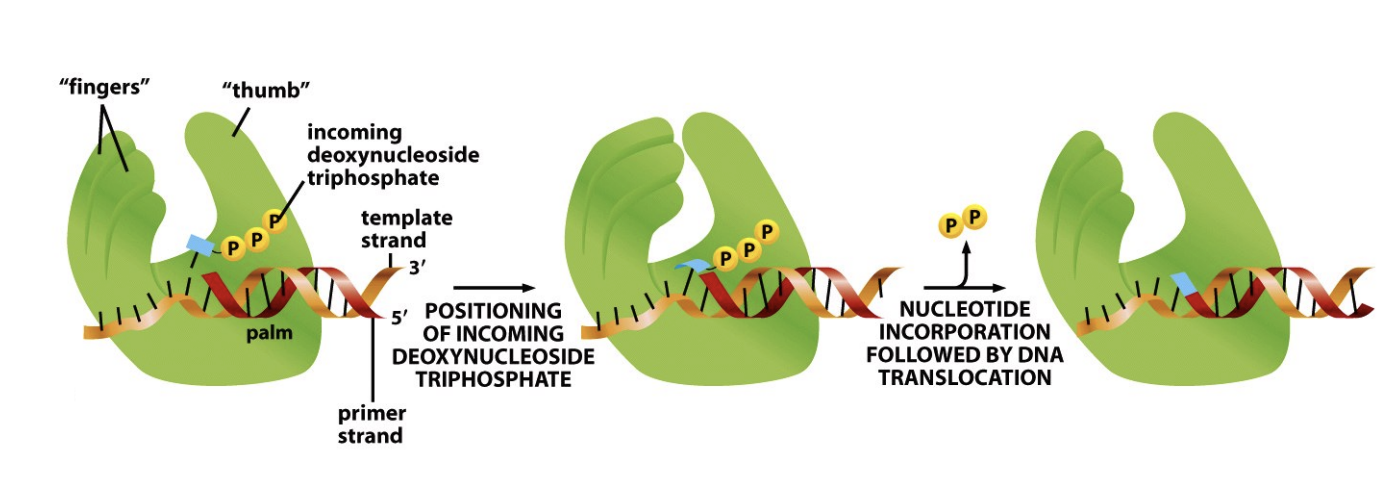
How does DNA polymerase acitivty work: hand fist anaology
Open lose grip hand
with incoming deoxynucleoside triphosphate nearby
closed fist
positions the triphosphate
nucleotide incorporated
open again
phosphates leave
perhaps need to look at how detailed the text book is of this process for more info!
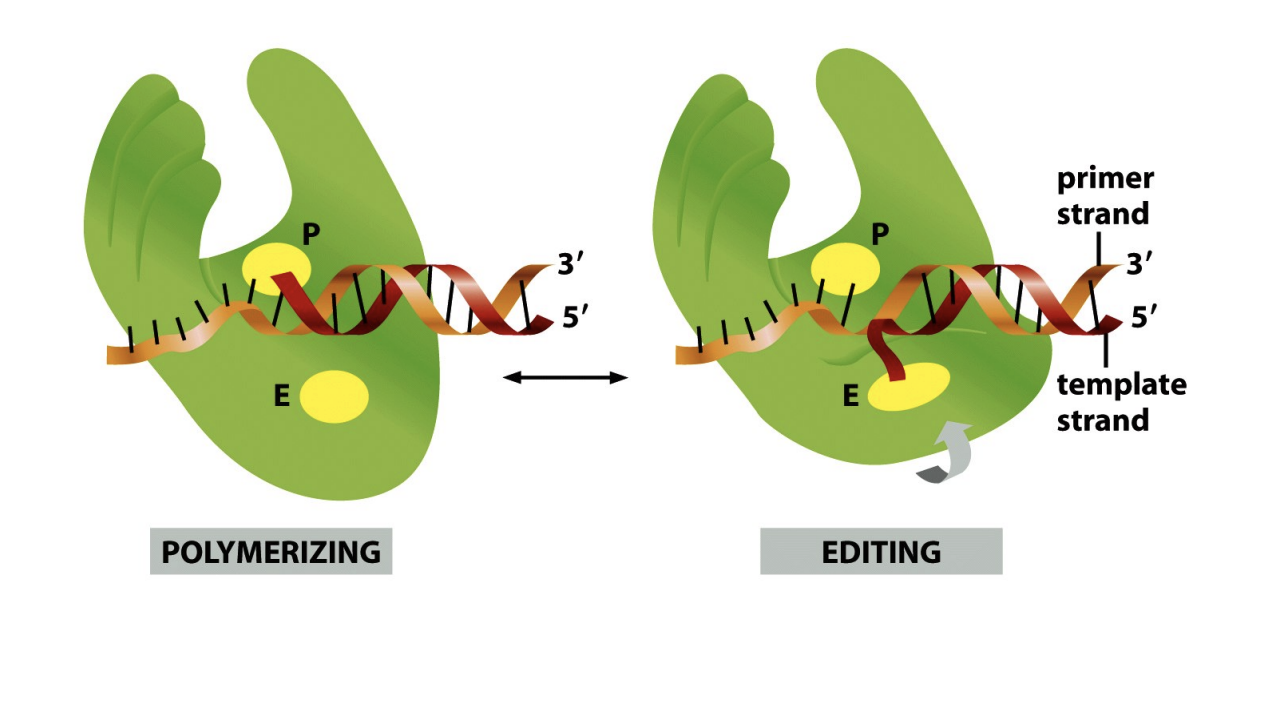
But, DNA polymerases make mistakes, they are corrected by…
Proof-reading exonuclease activity
by mismatch repair systems
can tell that there is a mismatch becase the DNA doesn’t bind as well and is bulky
diagram shows→ the polymerase shuffles back along the DNA to re-do the mistake it just made
Error rates of the pol
pol alpha is the worst
pol gamma or epsilon
pol gamma or epsilon
Why need low error rate in eukaroytes?
multicellular organism
other cells rely on these cells
→ can get benefits from duplicated genes that are mutated and do not have such a detrimental effect
in bacteria etc→ mutations are actually fab

Replication factors: other crucial proteins of the eukaryotic DNA replication fork include:
DNA helicases→ unwind the two DNA strands, generate ssDNA templates
RPA→ single strand binding protein, stabilities the unwound strands, recruits pol alpha/primase
PCNA→ sliding clamp, binds to pol gamma and epsilon, Fen-1 and others
RFC→ loads and unloads PCNA
Fen-1→ flap endonnuclease, removes sort primers
Dna2→ endonuclease, removes long primer flaps
DNA ligase I→ joins Okazaki fragments
DNA topoisomerases→ release superhelical stress
there is co-opertation of replication fork proteins with DNA polymerases

Please note:
my notes from BoC are so much better than this
On top of this…recent proteomic analysis of isolated DNA replication fork complexes have identified…
large amounts of additional proteins which play a role in
maintaining replication fork stability
facilitating replication of damaged DNA
replication of chromatin templates
DNA synthesis at DNA replication forks: concerted action of these core replication proteins during DNA strand synthesis in eukaryotes
INitiation and elongation of DNA strand synthesis
→ applies to both leading and lagging strand
Maturation of Okasaki fragments (lagging strand)
INitiation and elongation of DNA strand synthesis
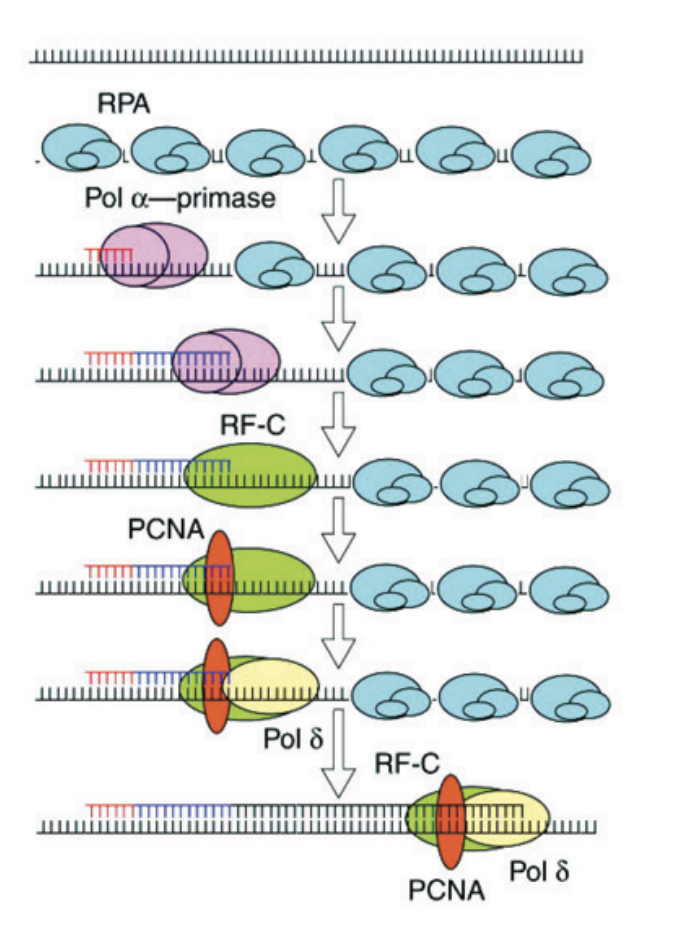
Replication Protein A (RPA)→ cover the single-stranded DNA
used to protect the DNA from endonucleases
AND signals for polymerase alpha
in open hand atm
Pol alpha PRIMES the DNA
Pol alpha primes and extends the leading strand
but then BIG pol delta/epison needs to get onto and stay on DNA, how?
Replication factor-C RF-C→ detect end
displaces alpha polymerase
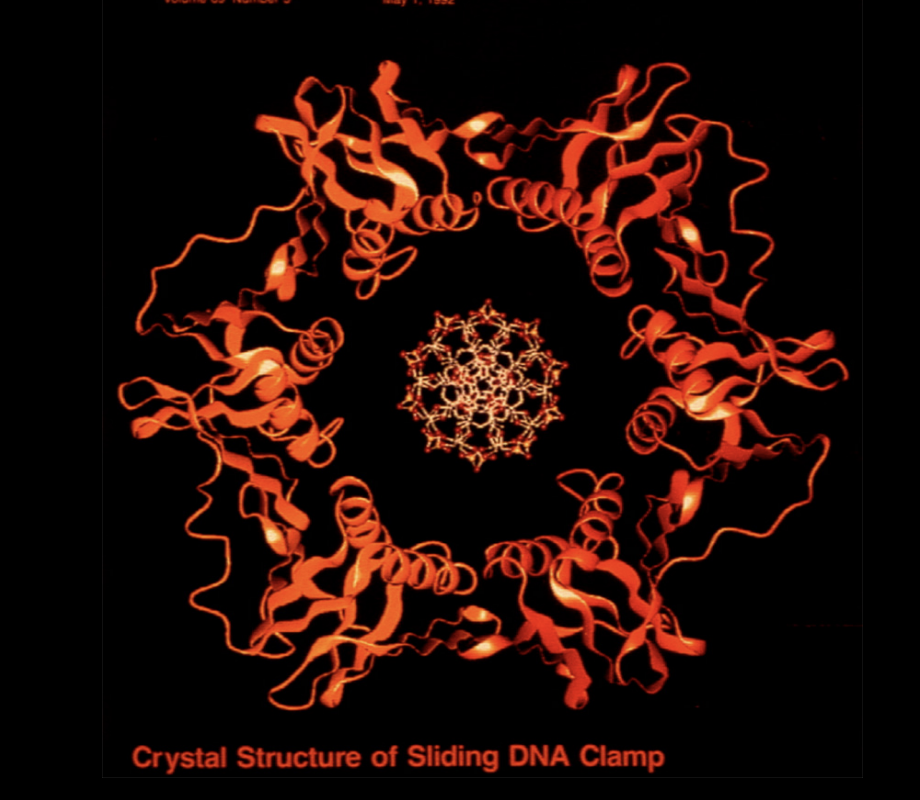
Recruits PCNA→ acts as sliding clamp (forms a circle around DNA to pull it through
binds to epsilon/alpha
pol d and e now on the DNA
Thousands of base pairs can now easily be polymerased
Crystal structure of sliding clamp
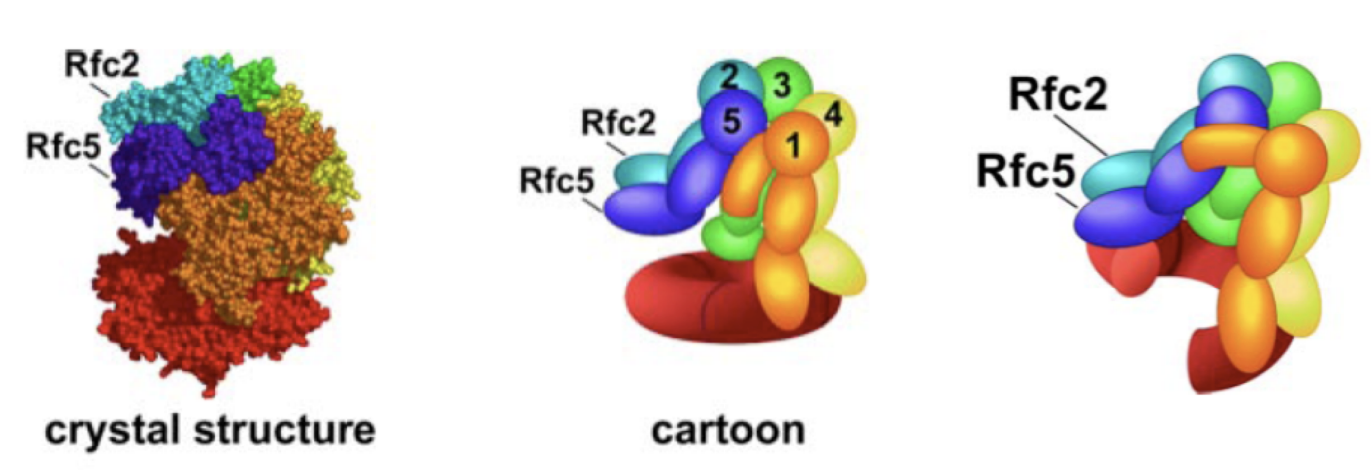
How does RF-C work to recuit PCNA
opens the PCNA ring like a spring washer
→ kinda slides open
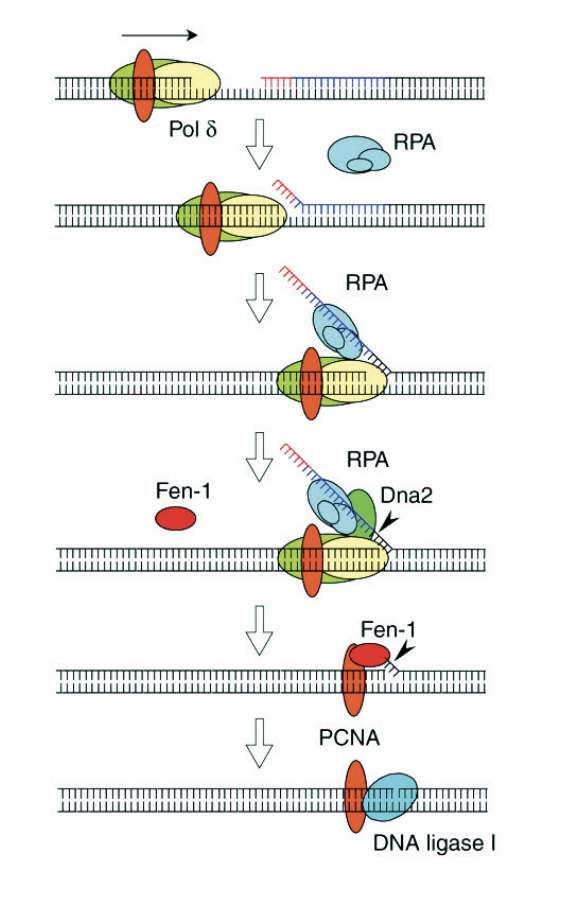
Maturation of Okasaki fragments (lagging strand)
pol delta ploughs through
gets to the double stranded ends from the other lagging strand/fragment
RPA is recuited→ stabilises the singlge stranded strands that is on this flap
This reuicts Dna2→ endonucleauses
cuts the DNA NON PRECISE
RPA leaves and Fen-1 is recruited→ flap endonuclease
this removes primers PRECISE
Fen-1 leaves
PCNA remains and recruits DNA ligase 1 to close the gap→ binds phosphate back bone together
To establish a DNA replication fork…
both leading and lagging strand synthesis are coupled
the lagging strand is looped back to obtain co-linearity
→ trombone model
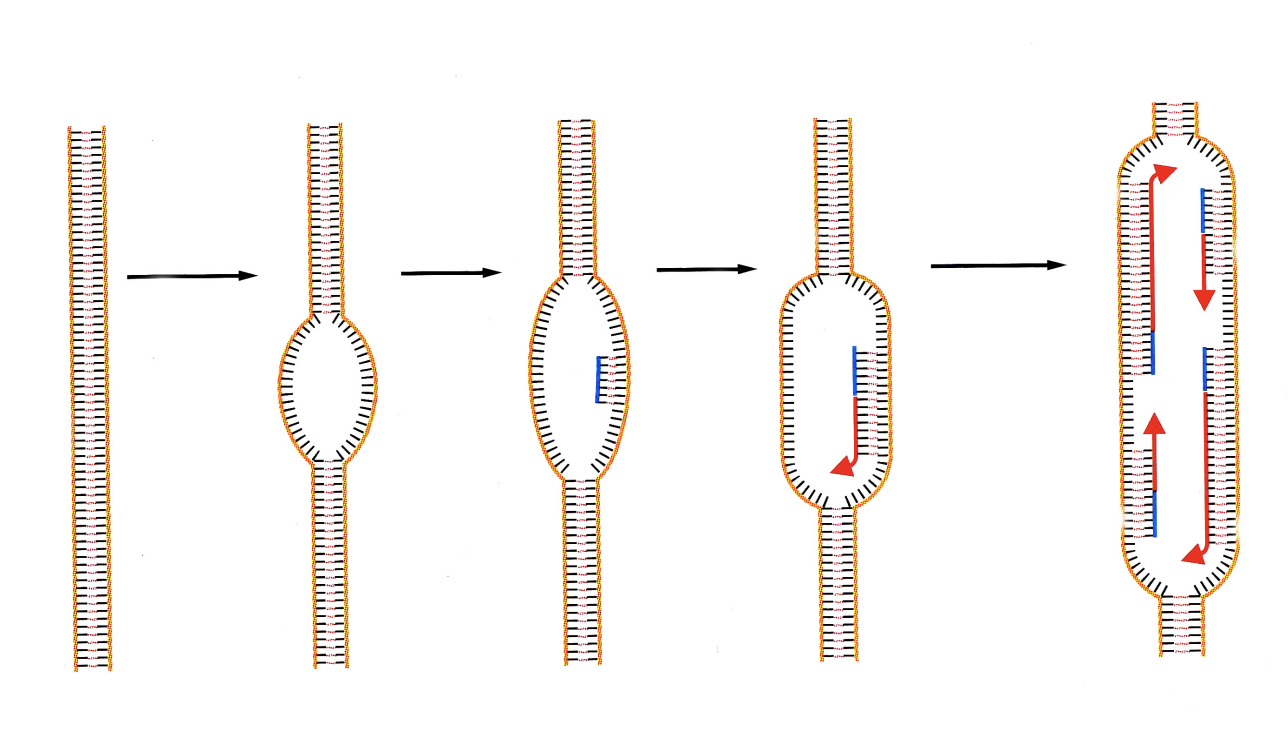
DNA replication is localised in areas
the DNA itself must be moved by this enzymatic activity
the DNA itself doen’t move by itself obvs
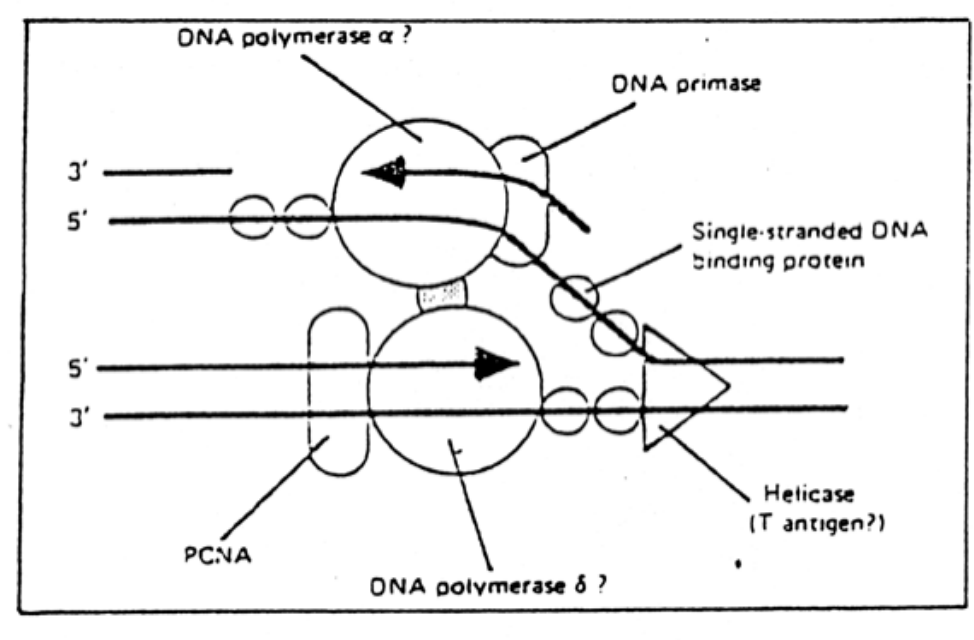
Historic 2D model of a replication fork (SV40)
however
→ this causes circular movement!
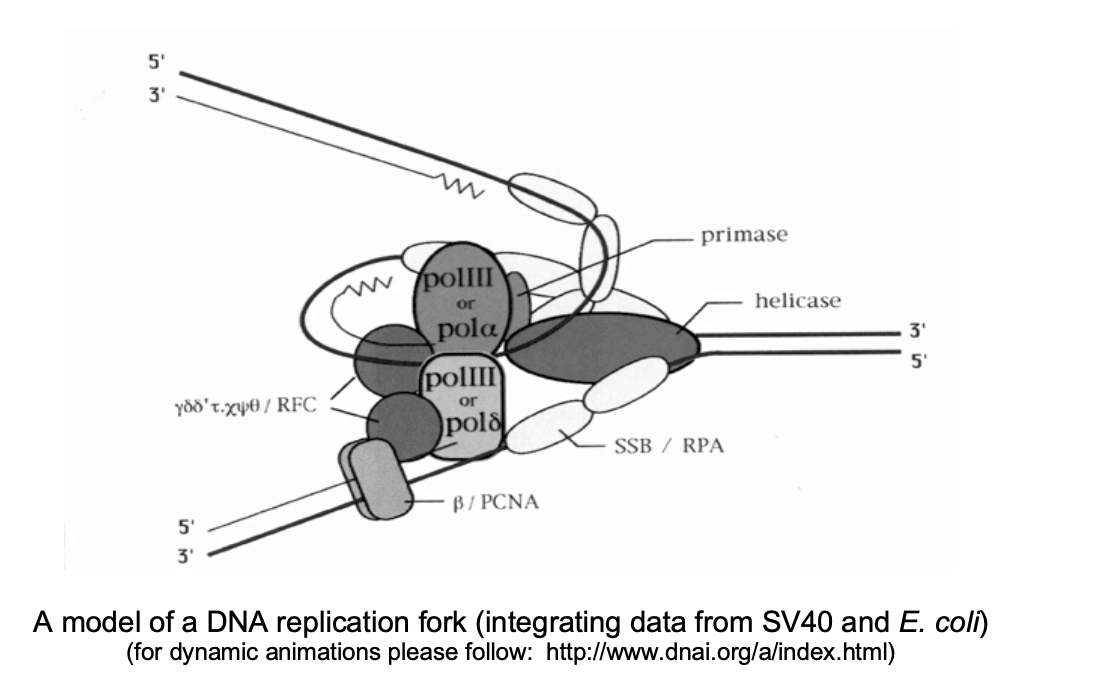
The proteins of the replication fork thus form…
a complex ‘molecular machine’
→ co-linear leading and lagging strand synthesis is now enabled with this formation
DNA topoisomerases: The immense length of DNA in the nucleus generates topological problems…
Winding
Average human chromosome (150Mbp)
DNA strands wind around each other (1.4 ×107 times)
These turns must be removed during replication
Supercoils
separation of DNA strands during transciption and DNA replication generates positive supercoils
ahead of the moving polymerase complexes
would eventually pprevent further elongation
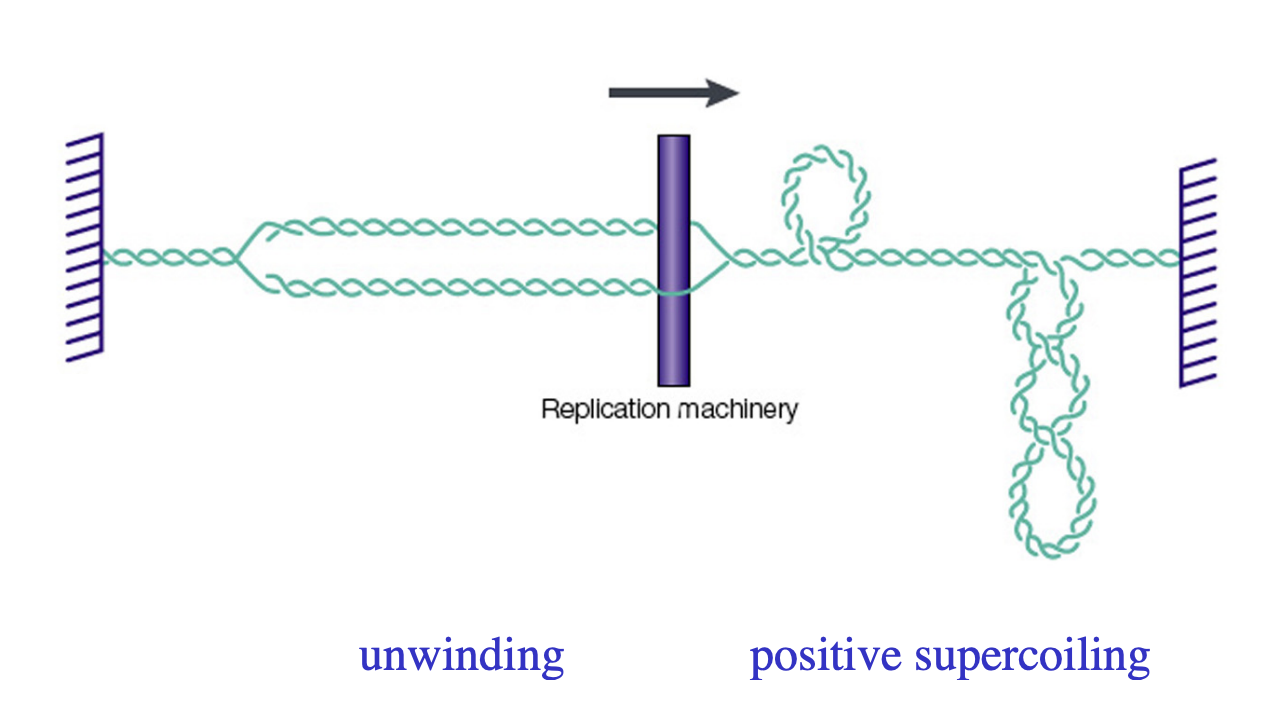
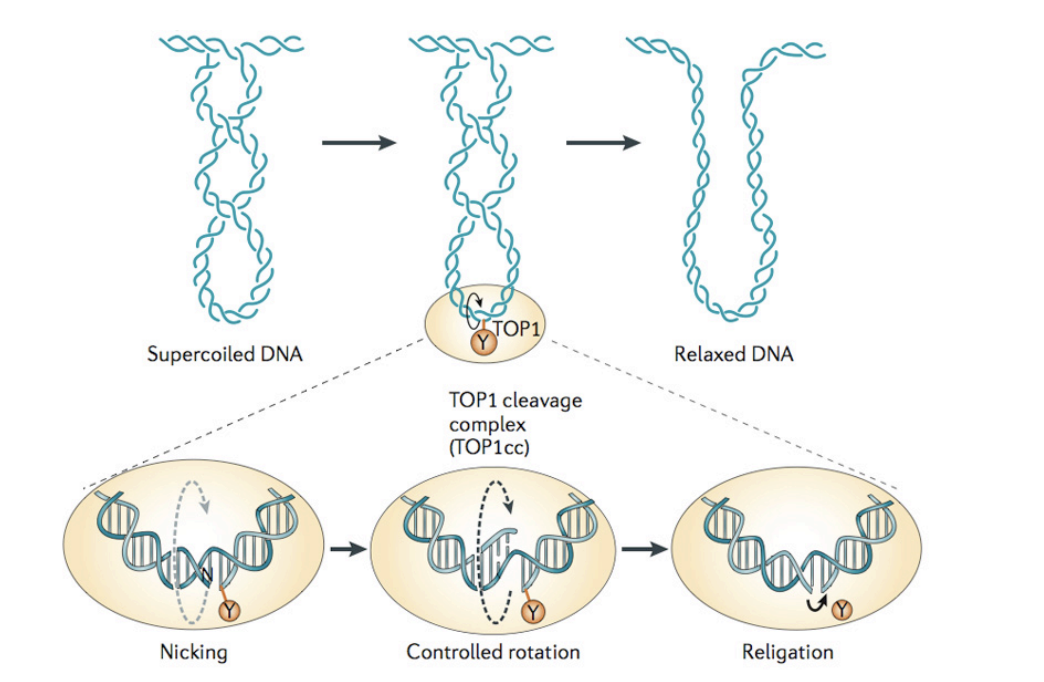
How do DNA topoisomerases resolve this issue
altering the number of times DNA strands wind around each other
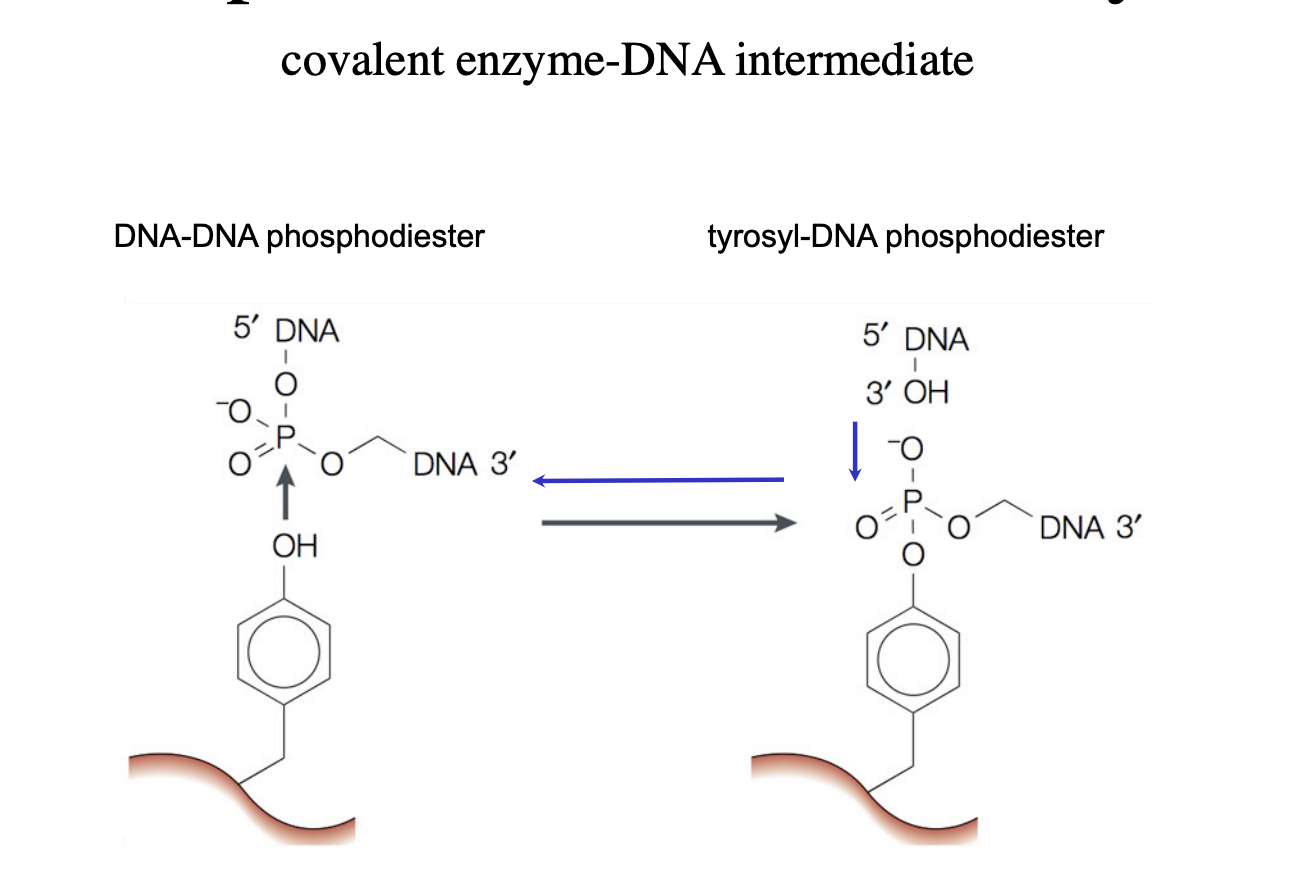
How does DNA topoisomerase I work?
nicks one strand of a DNA duplex
attaches a DNA phosphate group to a tyrosine residue in its active centre
covalently forming a new ester bond
Allows roatation of the free end of the cut strand around the uncut single-strand
seals the nick
breaks the ester bond of the DNA
with tyrosine
re-ligating the DNA without requiring ATP
these are trans-esterifications
This process can therefore…
Remove strain imposed on a molecule by local helix unwinding
as found in front of active DNA or RNA polymerases
Topoisomerase biochemistry
Problem when two forks meet
Catenation
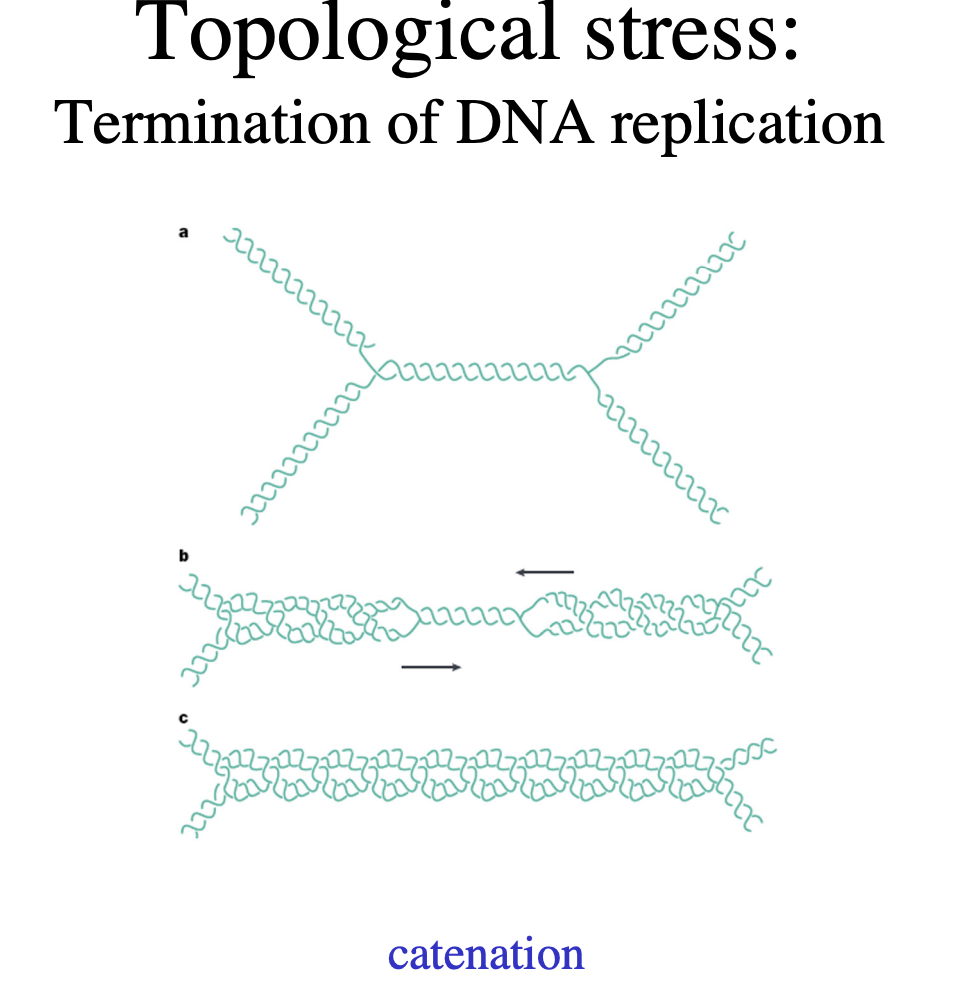
Solution to this
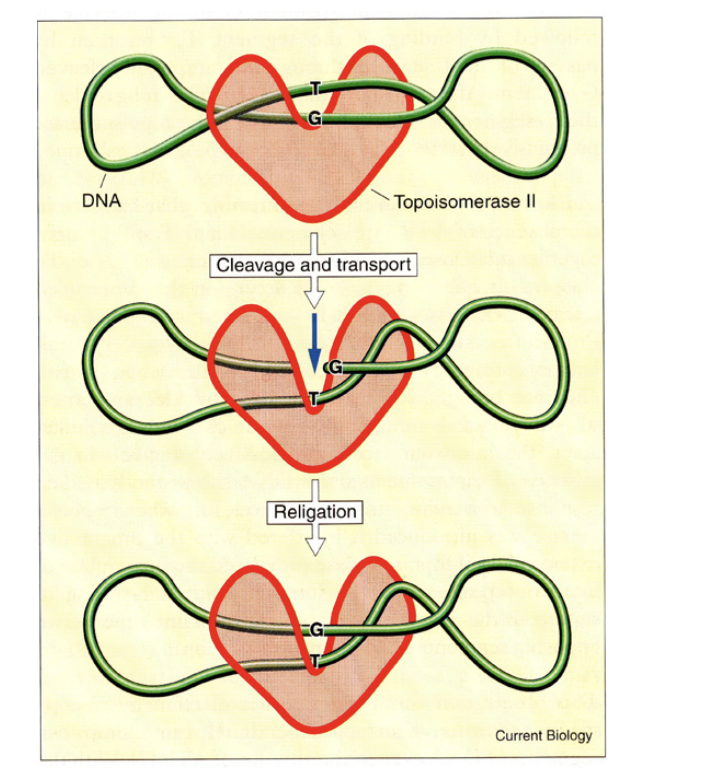
Topoisomerase II
→ two subunits, two DNA strands cut
HOw does DNA topoisomerase II work?
cuts both strand
bridges the gap
allowing other regions of DNA duplex to pass through before
resealing, removing supercoils from the DNA
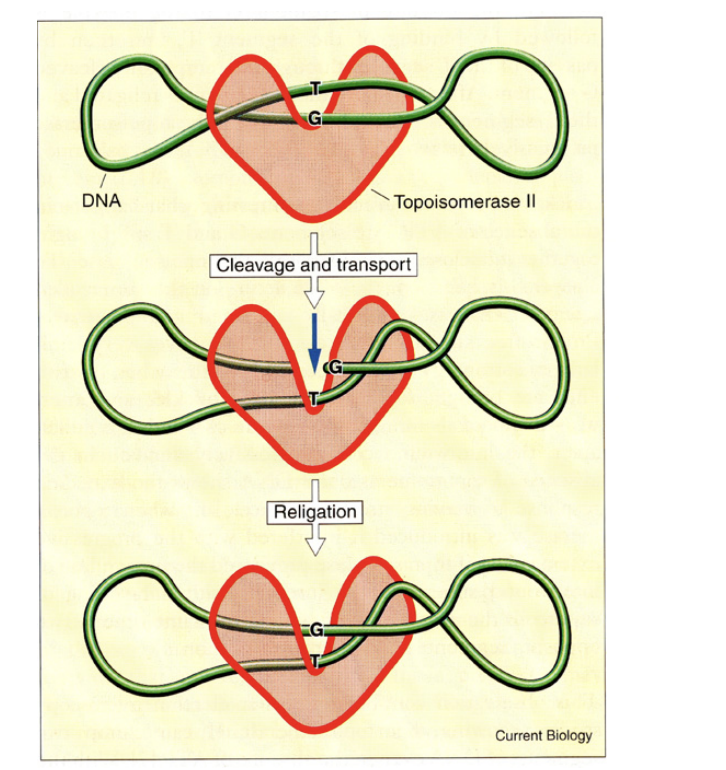
This enzyme can also…
Separate interlocked DNA rings (concatemers or catenanes)
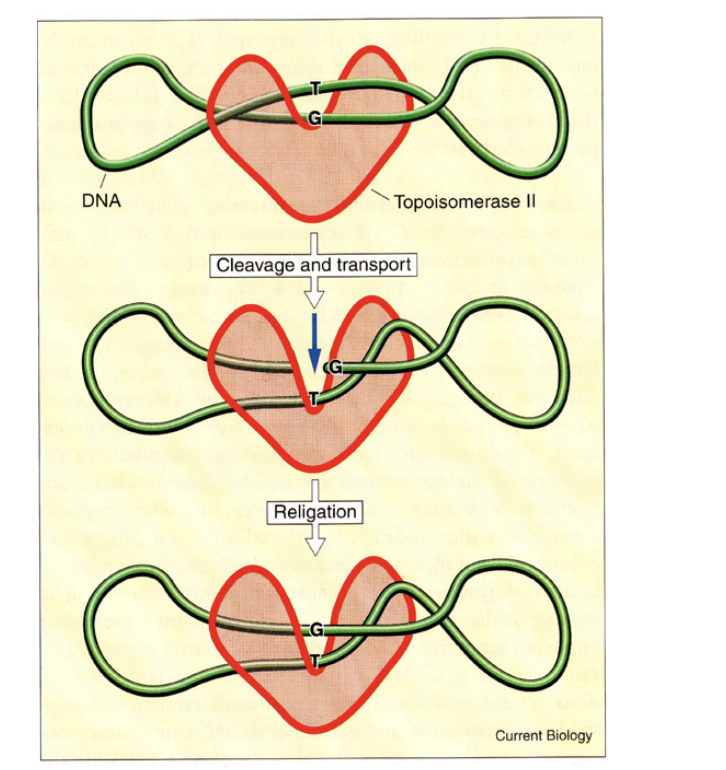
This property is essential in
the final stages of DNA replication
and during mitosis
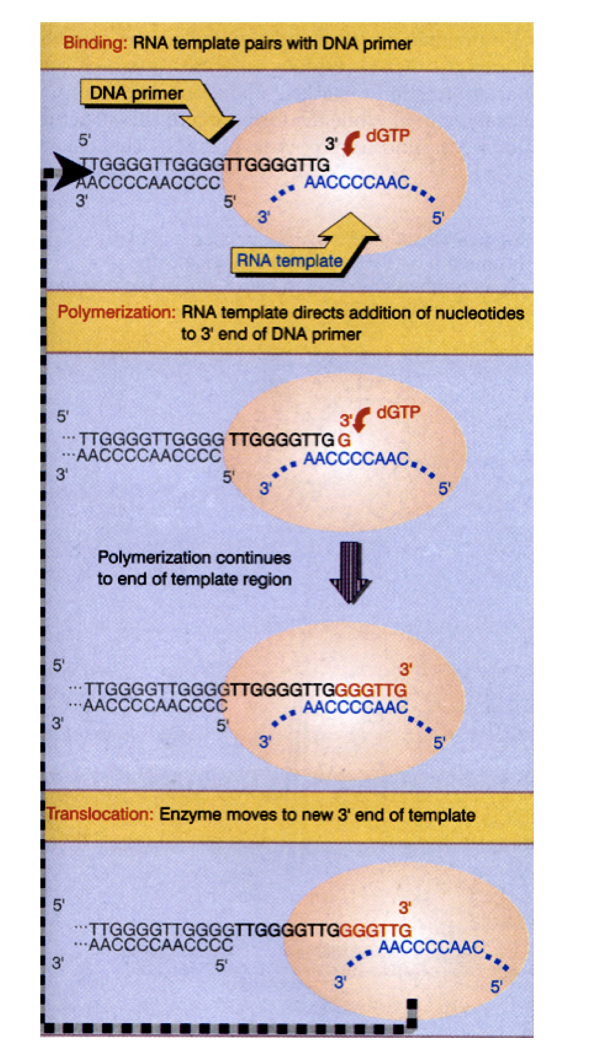
Replication of telomeres: termination problem
mechanism of co-ordinated leading and lagging strand synthesis
leads to loss of DNA at the linear ends of the chromosomes→ telomers
shorted after each cycle
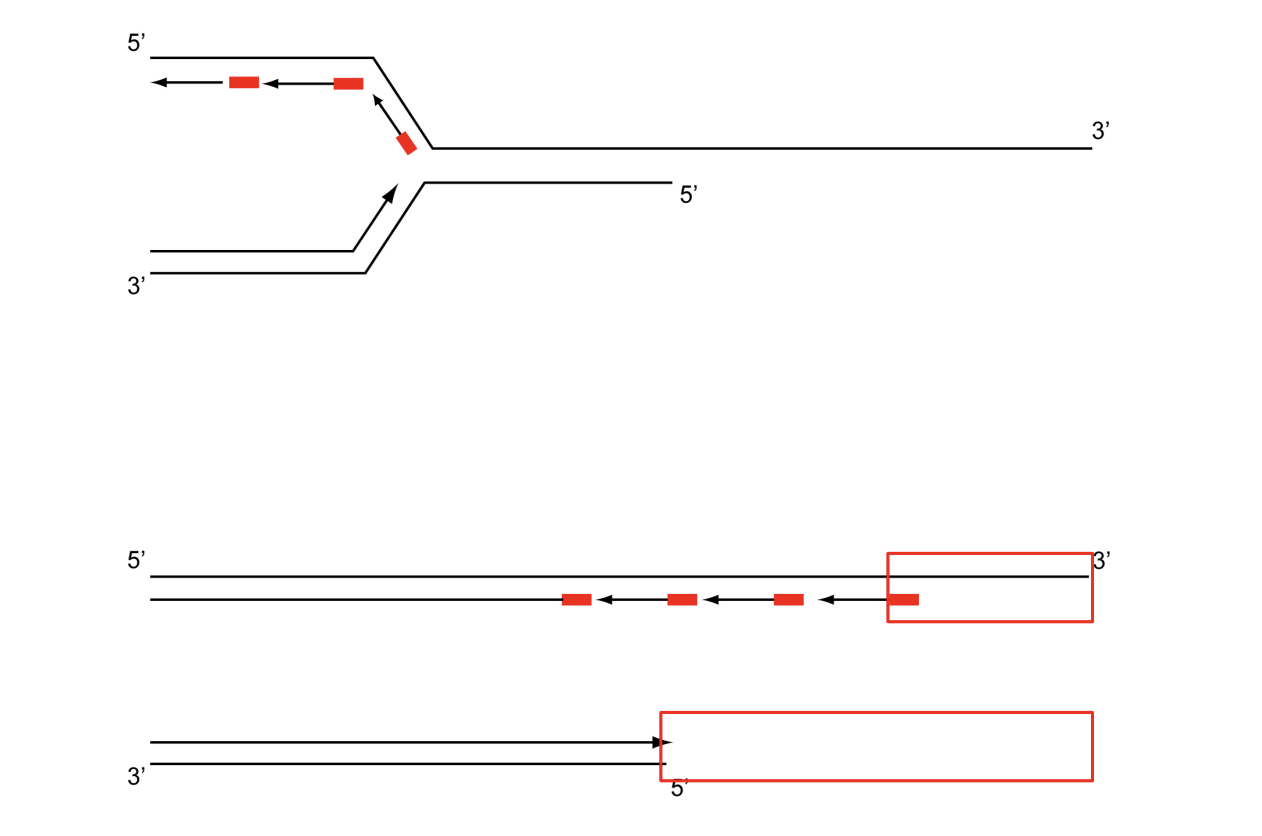
How is this loss counteracted?
Enzyme telomerase can elongate the ends by
synthesising and adding new telomere repeats onto the ends
using own RNA template
from diagram:
Binding→ RNA template pairs with DNA primer
Polymerisation→ RNA template directs addition of nucleotides to 3’ end of DNA primer
Translocation→ enzyme moves to new 3’ end of template
What cells have telomerase
germ cells
cancer cells
immortalised cells→ HeLa