Secondary Structure
1/17
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
18 Terms
The distance between hydrogen bond donor and acceptor is within ... Å and the angle formed by the donor, hydrogen, and acceptor is around ... degrees.
3.0 and 180
Hydrogen bond distances are ~ 2 Å. Covalent bonds are ~ 1 Å. The distance between the hydrogen bond donor (i.e. N or O of N-H or O-H) and acceptor is ~ 3 Å.
The strongest hydrogen bonds have the hydrogen-bond donor, the hydrogen atom, and the hydrogen-bond acceptor along a straight line.
Hydrogen bonds in β-sheets are …
Roughly perpendicular to the polypeptide chain direction
β-sheets contain extended stretches of polypeptide chain with hydrogen bonds between neighbouring strands between C=O and N-H groups. These hydrogen bonds are roughly perpendicular to the polypeptide chain direction.
Hydrogen bonds have bond distances ranging from ~ 2 Å.
In an α-helix, the hydrogen bonds …
Are roughly parallel to the axis of the helix
The α-helix is stabilised by hydrogen bonds between C=O and N-H groups that are four residues along.
They are roughly parallel to the axis of the helix.
In an α-helix, the R groups on the amino acid residues …
Are found on the outside of the helix spiral
In an α-helix, the side-chains (R-groups) project outwards from the α-helix as it rises; thus steric interference with the backbone or with other side-chains is avoided.
A favourable charge-charge interaction between R groups in an α-helix is expected to occur when the interacting side chains are separated by …
Three-four residues
This is due to a rise of 1.5 Å and a rotation of 100° between each residue, which results in 3.6 residues per turn of the helix.
This means that residues x and x + 3 / 4 are spatially quite close, they are found almost directly above or below each other.
Charge-charge interactions between the side chains of these residues are possible.
An α-helix with 11 residues completes three turns. The length of this helix is about …
15 Å
Each residue is related to the next one by a rise of 1.5 Å along the axis and a rotation of 100 degrees, which gives 3.6 amino acid residues per turn. The pitch of the α-helix, which is equal to the product of the translation (1.5 Å) and the number of residues per turn (3.6), is 5.4 Å.
An α-helix with 11 residues and three turns is ~ 16 Å.
Which amino acids disrupt an a-helix?
Pro and Gly
Pro (structurally rigid) and Gly (too small) are known as "helix breakers" because they disrupt the α-helical backbone conformation.

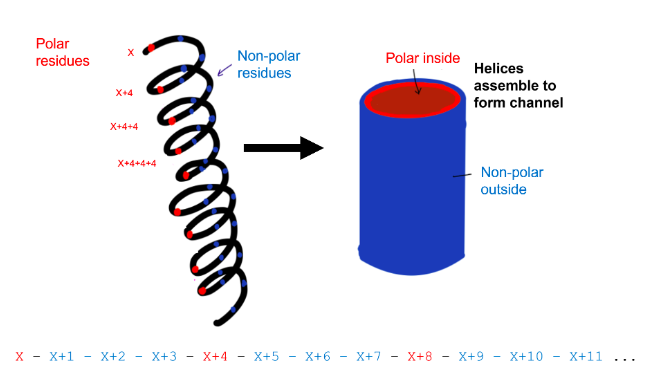
Amphipathic α-helical regions of proteins can cluster together to form an ion channel through a plasma membrane (e.g. the potassium channel). Bearing in mind which amino acids are compatible with forming an α-helix and the average number of amino acid residues per helical turn, which of the following polypeptides would be most likely to form an amphipathic helix in an ion channel?
Helix 2
First, find a sequence that can form an α-helix. Remember, Pro and Gly are helix breakers. Sequence 1 contains multiple Gly residues, sequence 3 contains multiple Pro residues. Both sequences are unlikely to form α-helices. This leaves us with sequences 2 and 4, both contain one Gly residue.
Then, we need to choose a sequence that is compatible with an amphipathic α-helix in an ion channel. The helix should have polar and non-polar residues as one side of the helix needs to be polar (the inside of the channel, where potassium travels through the membrane) and the other side needs to be non-polar (the outside of the channel, in contact with the lipids of the membrane). The amino acids in sequence 2 have the following pattern: x and x+4 and x+4+4 and ... are polar, while the rest is non-polar. As one turn of an α-helix contains 3.6 residues, this pattern fits with a sequence compatible with an amphipathic α-helix. See the figure by Nuo Xu, class of 2020 for clarification.
Sequence 4 does not fit the correct pattern for an amphipathic α-helix. Its N-terminal part is non-polar, its C-terminal part is polar.
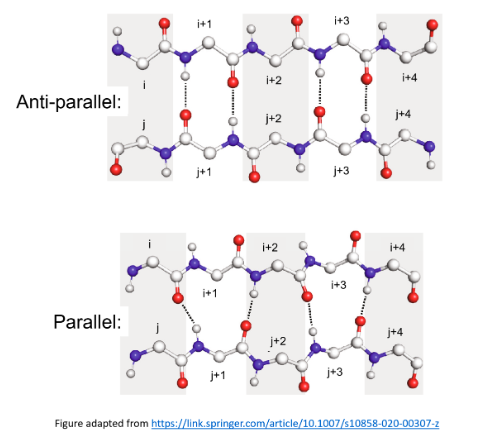
The major reason that antiparallel β-sheets are more stable than parallel β-sheets is that the latter …
Have weaker hydrogen bonds laterally between adjacent strands
The figure below displays antiparallel and parallel sheets, each consisting of two strands, and each strand has 5 residues. There are 4 hydrogen bonds for both sheets.
Parallel sheets have weaker hydrogen bonds laterally between adjacent strands as the hydrogen bonds do not have the hydrogen-bond donor, the hydrogen atom, and the hydrogen-bond acceptor along a straight line.
Amino acid residues commonly found in the middle of a β-turn are …
Pro and Gly
Certain amino acids (i.e. Pro, Asn and Gly) are observed more frequently then others in β-turns, reflecting the properties of their R-group and the φ and ψ angles they can adopt.
β-sheet
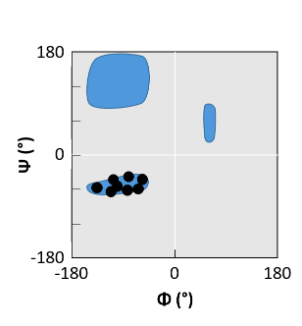
helical peptide
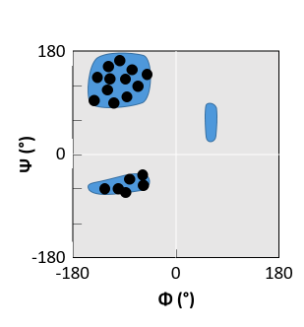
βαβ
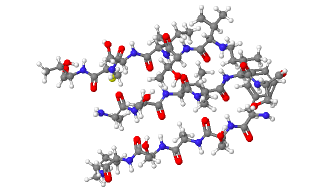
Investigate the β-sheet shown.
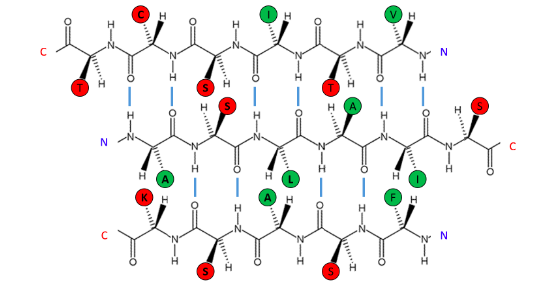
The following β-sheet is most likely to be ...
On the surface of the protein
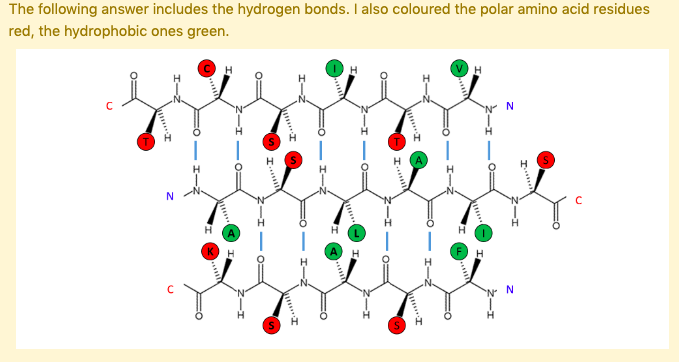
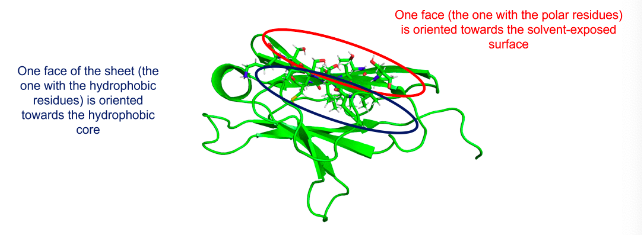
Hydrophobic amino acids are concentrated on one side of the β-sheet, polar or hydrophilic amino acids on the other. This arrangement is common in β-sheets on the surface of the protein, where one face of the sheet (the one with the hydrophobic residues) is oriented towards the hydrophobic core and one face (the one with the polar residues) is oriented toward the solvent-exposed surface.
How does this look like?
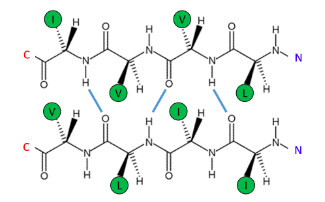
The following β-sheet is most likely to be ...
Buried in the interior of the protein
Hydrophobic amino acids are concentrated on both sides of the β-sheet. This arrangement is common in β-sheets within globular proteins, where both sides are oriented towards other hydrophobic amino acids.
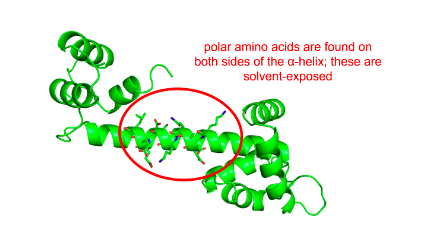
Investigate the a-helix shown.
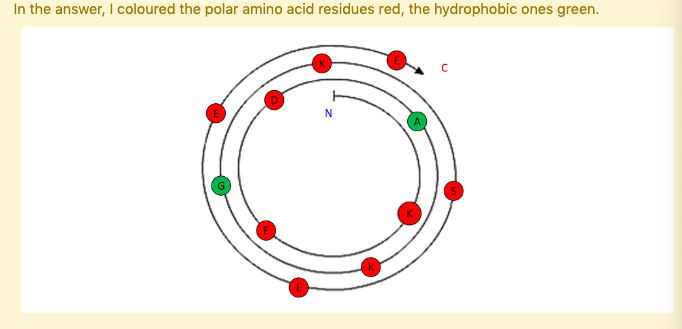
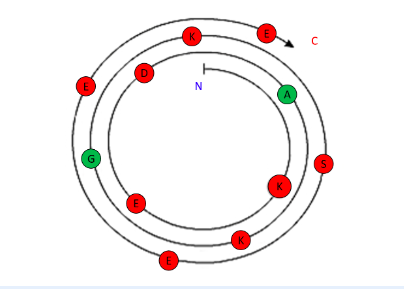
The following α-helix is most likely to be ...
Fully-solvent exposed
Polar amino acids are found on both sides of the α-helix. These polar residues are most likely exposed to the solvent.
How does this look like?