Part 2.2: Transport to Organelles and Nucleus
1/49
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
50 Terms
Mitochondrial Import: Background
most mitochondrial proteins are encoded in the nucleus and synthesized in the cytosol
precursor proteins are synthesized on cytosolic ribosomes; chaperons like Hsc70 bind to them and keep them unfolded
only unfolded protein can thread thru the import pores
mitochondrial precursor proteins have an N-terminal targeting signal that is different from ER signal sequences
typically amphipathic ⍺-helix
recognition depends on specific protein interactions with mitochondrial receptors, not just hydrophobicity
ATP hydrolysis helps drive import into the matrix
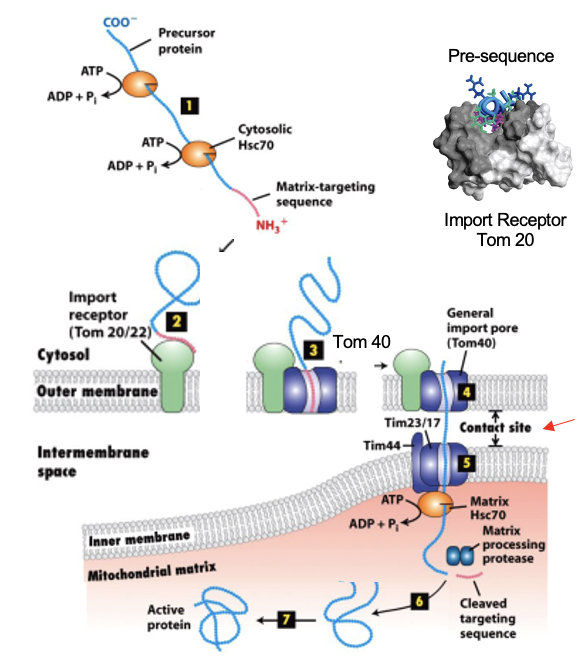
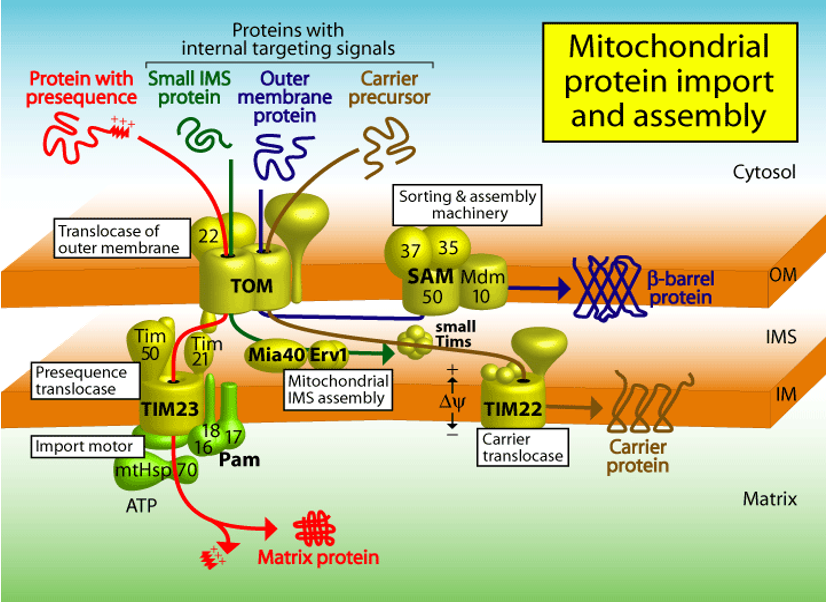
Mitochondrial Transport: TOM
Translocator of Outer Membrane Complex
TOM20: receptor recognizing the targeting signal (import receptor)
TOM40: the actual pore that allows the unfolded protein to cross the outer membrane
After TOM20 binds the signal, the precursor is handed off to TOM40
Mitochondrial Transport: TIM
Translocator of Inner Membrane Complex
works with TOM at contact sites where the inner and outer membranes are close
translocates proteins across the inner membrane into the matrix
Mitochondrial Transport: Driving Forces
matrix chaperones (eg. Hsc70) bind to the incoming protein inside the matrix and use ATP hydrolysis to pull it through
proton motive force (PMF) across the inner membrane also helps drive import, especially for inner membrane proteins
the intermembrane space is more positively charged relative to he matrix, the PMF pulls + charged portions of precursors inward
Mitochondrial Matrix: Additional Steps
after import, the N-terminal targeting signal is cleaved by a peptidase
final folding is assisted by matrix chaperones
Mitochondrial Matrix: Steps
protein synthesized in cytosol → bound by cytosolic chaperones
N-terminal mitochondrial signal recognized by TOM 20 receptor
protein enters TOM40 pore
protein passes TIM complex at inner membrane contact sites (only for matrix proteins)
matrix chaperones + ATP pull protein in; PMF may assist
signal peptide removed → protein folds into functional form
Mitochondrial Import: Figure
Mitocondrial Import: Porins
the TOM complex cannot alone integrate β-barrel porins into the lipid bilayer from outside
porins are first transported into the intermembrane space, binding specialized chaperones
porins then bind to the SAM complex in the outer membrane, which inserts them into the outer membrane
the central subunits of the SAM complex are homologous to a bacterial outer membrane protein that helps insert β-barrel proteins (eg. BAM in bacteria)
Chloroplast Import: Background
protein import into chloroplasts happens post-translationally (proteins are synthesized
resembles transport into mitochondria
proteins remain unfolded, assisted by cytosolic chaperones, until they reach the chloroplast
signal sequences for import into chloroplasts resemble those for mitochondria
Chloroplast Import
TOC (translocon at outer chloroplast membrane)
TIC (translocon at inner chloroplast membrane)
these are homologous to mitochondrial TOM and TIM complexes
the chloroplast-targeting signal sequence at the N-terminus directs the protein to the TOC receptor
Chloroplast Import: Thylakoid
chloroplasts have an additional membrane-enclosed compartment, the thylakoid
the photosynthetic system is located in the thylakoid membrane
proteins destined for the thylakoid need 2 targeting signals
chloroplast signal sequence (for import into stroma)
thylakoid signal sequence (for further transport across thylakoid membrane)
Chloroplast Import: Two-Step Transport Pathway
precursor proteins cross the outer and inner envelope membranes thru the TOC and TIC complexes at contact sites
once inside the chloroplast-targeting signal is cleaved off
the remaining thylakoid signal sequence directs the protein to the thylakoid membrane or lumen
via a similar system to SRP and SecYEG of bacteria
Peroxisomes
all eukaryotic cells have peroxisomes
sites of oxygen utilization, β-oxidation of fatty acids into acetyl-CoA, ethanol oxidation to acetaldehyde
surrounded by a single membrane
acquire proteins from the cytosol, including oxidative enzymes, such as catalase and urate oxidase (present at high concentrations in the peroxisome)
enzymes are not made inside
Peroxisomal Targeting Signal (PTS1)
the sequence (Ser, Lys, Leu) at the C-terminus of peroxisomal proteins is the import signal
not cleaved off after import
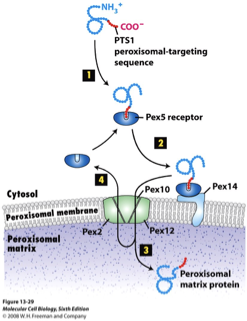
Peroxisome Protein Import: Peroxins
at least 23 PEX proteins mediate peroxisomal import
import is post-translational: proteins are fully folded in cytosol before import
PEX5: the cytosolic receptor for the SKL signal, binds SKL-containing proteins in the cytosol, docks at the peroxisome membrane
after delivering cargo, PEX5 is ubiquitinated, extracted and reuse
PEX13 & PEX14: form the docking complex on the peroxisome membrane, accept PEX5+cargo
PEX2, PEX10, PEX12: required for the translocation of cargo and for recycling PEX5 back to the cytosol
Peroximal Protein Import FIGURE
Peroxisomal Import: Zellweger Syndrome
caused by a defect in importing proteins into peroxisomes and peroxin function
individuals have cells with empty peroxisomes, leading to severe brain abnormalities and die soon after birth
Primary Hyperoxaluria
protein targeting is essential, proteins must be delivered to exactly the right organelle to function
Normal situation: the enzyme alanine:glyoxylate aminotransferase (AGT) normally needs to be in the peroxisome
contains a PTS1 at its C-terminus
in peroxisomes, AGT convert glyoxylate → glycine, preventing toxic buildup
Disease situation: PTS1 signal is altered, now interpreted as mitochondrial targeting signal
AGT cannot access glyoxylate, glyoxylate accumulates and converted to oxalate
oxalate forms insoluble calcium oxalate crystals leading to kidney stones in early childhood and renal failure if untreated
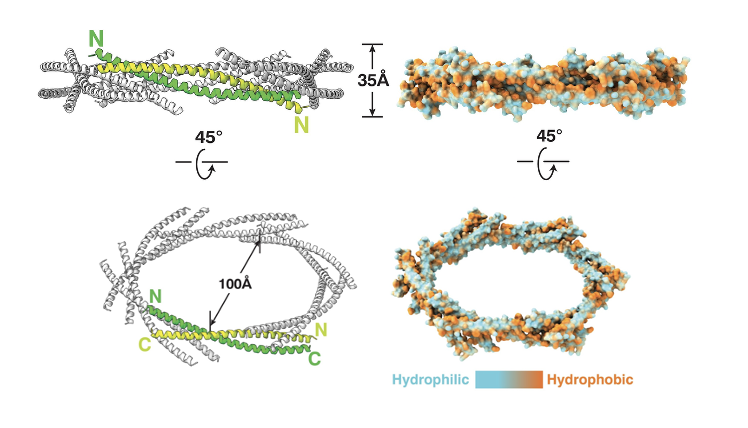
PEX13 Dodecamer Ring: Amphipathic Helix
PEX13 contains a long amphipathic helix, and 12 copies of this helix assemble into a ring-like pore in the peroxisomal membrane
the helices tilt and alternate orientation
the downstream TMS and SH3 domains also alternate around the ring (black dotted lines)
the ring creates a large central opening
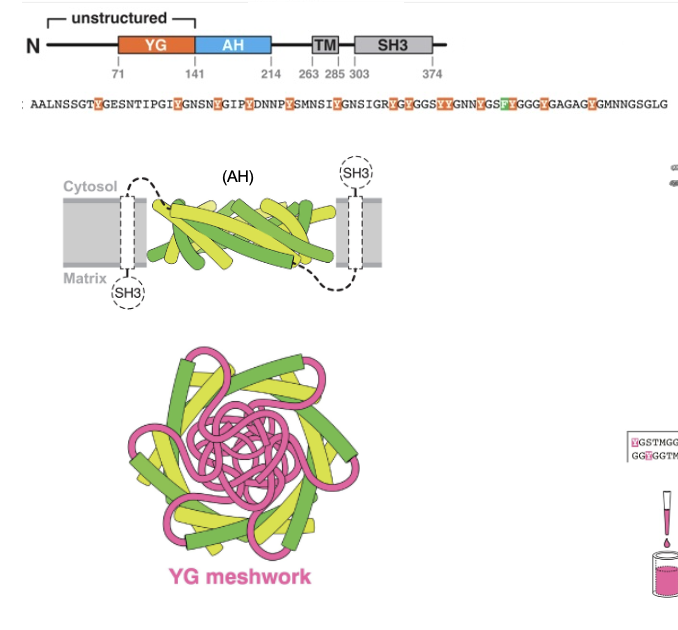
PEx13 Dodecamer Ring: Hydrophobicity/Hydrophilicity
hydrophobic residues of each helix face outward and interact with the lipid bilayer
hydrophilic residues face inward, forming a water-filled channel
PEx13 Dodecamer Ring: In the Membrane
PEX13 had a Tyr/Gly (YG) domain
inside, the YG domains from each subunit project inward to form a dense meshwork that acts as a selective barrier for protein import
nuclear pore-like hydrogel
the meshwork is held together by cohesive interactions between the Y residues of the YG domain
PEX5 partitions into this meshwork by transiently disrupting the cohesive interactions using its WxxF/Y motifs
these motifs temporarily disrupt the YG gel, allowing PEX5+cargo to pass thru the pore, then the gel reseals behind them
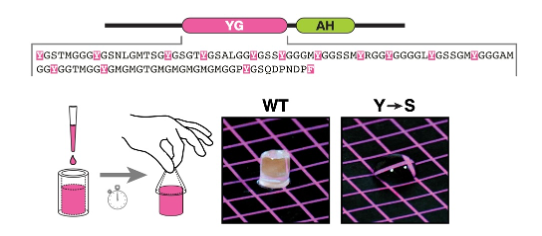
PEx13 Dodecamer Ring: Experimental Evidence
the purified YG domain of PEX13 forms a hydrogel
a concentrated solution (40 mg/mL) of the protein was pipetted into silicone tubing and allowed to gel, then squeezed out onto a colored surface and photographed
gelation was observed with the WT protein, but a mutant (Tyr→Ser) remained fluid
therefore, tyrosines are essential for hydrogel formation
PEX5 Receptor
cytosolic receptor that recognizes cargo that contains SKL signal and guides it through a docking complex on the peroxisomal membrane.
after releasing the proteins inside, the PEX5 receptor is recycled back to the cytoplasm to pick up more cargo
PEX5 is rich in aromatic residues (Trp, Phe, Tyr) in its N-terminal region, specifically within its repeated WxxF/Y motifs
allow for aromatic-aromatic (π-π) stacking interactions, which allow PEX5’s motifs to disrupt or slip between the Tyr’s in the YG domain of PEX13
allows to carry cargo thru the pore and exit again into the cytosol once cargo is released
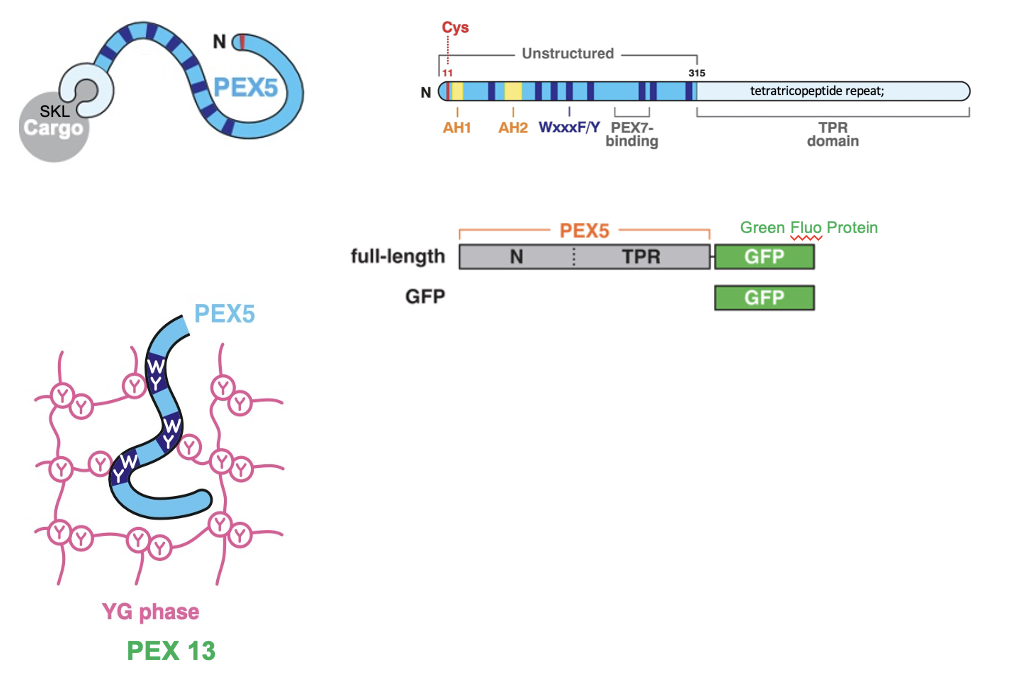
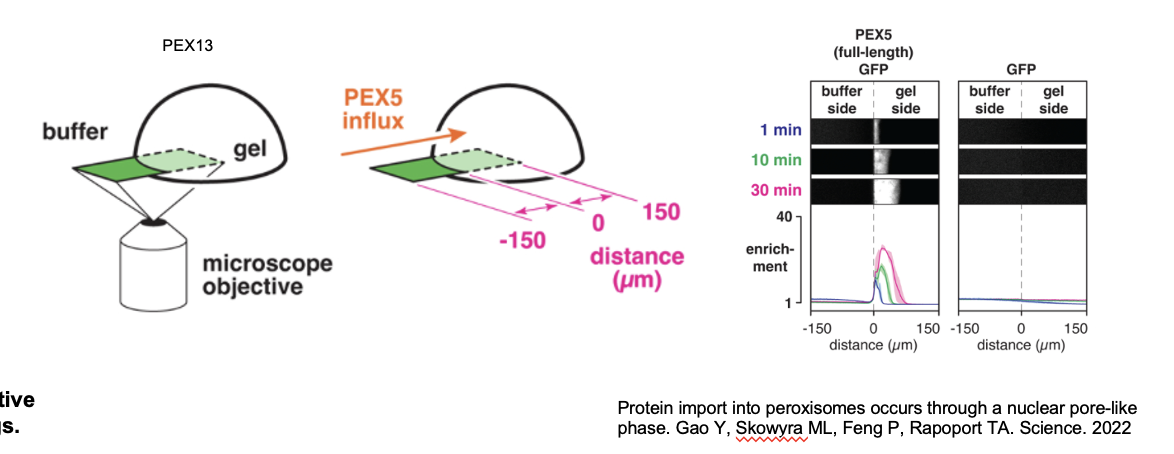
PEX5 Receptor: Experiment
YG-hydrogel droplets (40mg/ml) were prepared in glass-bottomed dishes; permeation of the gels by fluorescently labeled PEX5 or other proteins was images by point-scanning confocal microscopy
scheme depicting the PEX5 fragments that were fused to GFP. PEX5’s N-terminal region contains several WxxxF/Y motifs
YG-hydrogel droplets were bathed in buffer containing the indicated GFP-fusion proteins (or GFP alone), and the interface between the buffer and gel was imaged over time
shown at 3 selected time points; the fold enrichment of each protein, relative to buffer, across the imaged field is plotted below (mean ± the range of three experiments).
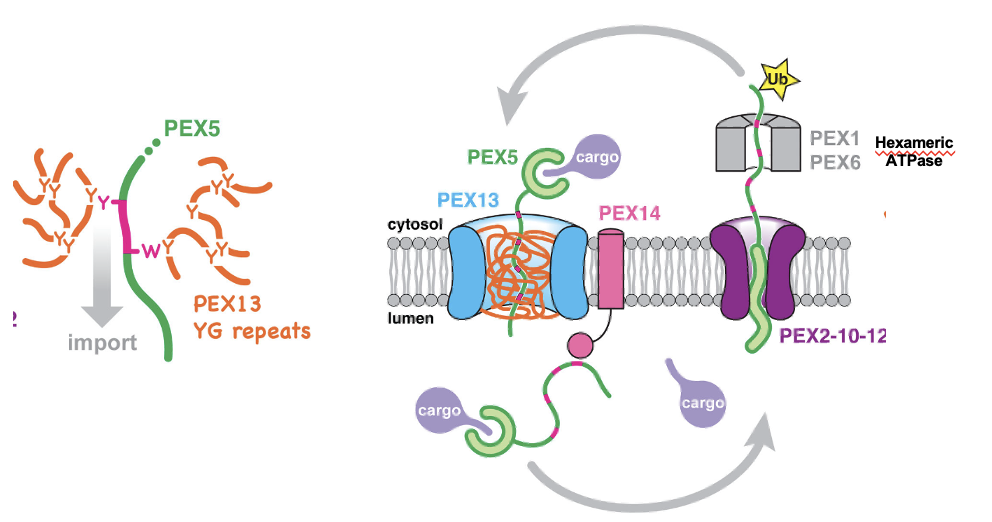
PEX Transport Cycle: Step 1
PEX5 recognizes and binds to cargo proteins in the cytosol that contain an SKL signal
PEX Transport Cycle: Step 2
Cargo-bound PEX5 is recruited to peroxisomes by a complex containing the membrane proteins PEX13 and PEX14
PEX Transport Cycle: Step 3
Cargo-bound PEX5 traverses the membrane through a conduit formed by multiple copies of PEX13. The conduit contains a dense meshwork formed from the PEX13 Tyr/Gly- rich domain, into which PEX5 partitions using its WxxF/Y motifs
PEX Transport Cycle: Step 4
PEX5 is drawn into the matrix by favourable lumenal interactions between the receptor WxxxF/Y motifs and PEX14 oligomers.
the lumenal interactions are energetically favorable, so they pull PEX5 deeper into the matrix, bring the cargo with it
PEX Transport Cycle: Step 5
PEX5 now needs to be removed from the matrix
PEX5 spools its flexible N-terminus into the cytosol through a pore in the PEX2-PEX10-PEX12 ubiquitin ligase complex, which then monoubiquitinates the receptor on a conserved cysteine
PEX Transport Cycle: Step 6
Monoubiquitinated PEX5 is pulled out of the matrix thru the ligase pore by the PEX1-PEX6 AAA ATPase, which unfolds the receptor and causes cargo to be stripped off inside the matrix
energy dependent step
PEX Transport Cycle: Step 7
PEX5 refolds in the cytosol and ubiquitin is removed by de-ubiquitinases resetting the receptor for another import cycle
PEX Transport Cycle Figure
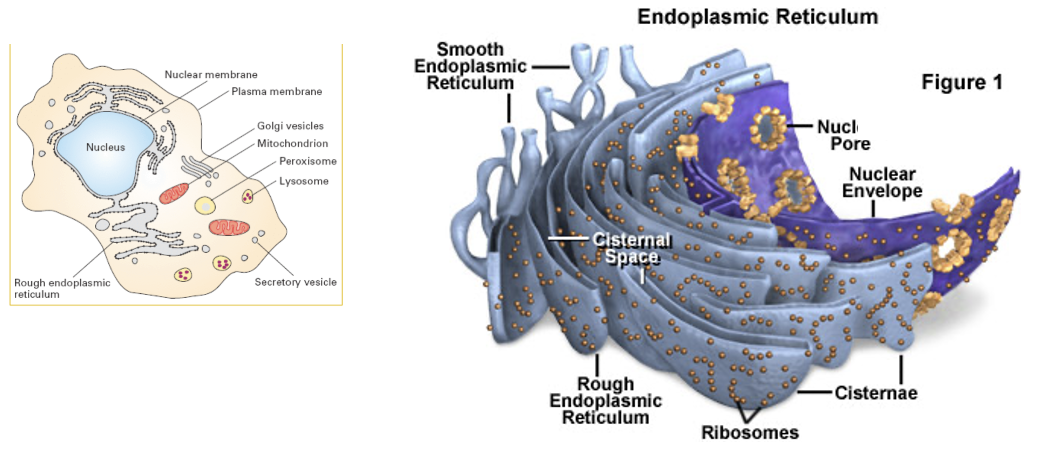
Nuclear Envelope
encloses the DNA and defines the nuclear compartment
the envelope consists of two membranes, penetrated by nuclear pore complexes
the inner membrane is an anchoring site for chromatin and for the nuclear lamina
the outer nuclear membrane is continuous with the membrane of the ER
protein traffic occurs continuously between the cytosol and the nucleus
many proteins synthesized in cytosol function in the nucleus (eg. histones, DNA and RNA polymerases, gene regulatory proteins and RNA processing proteins)
tRNAs and mRNAs synthesized in the nuclear compartment are exported to the cytosol
ER
organized into a netlike labyrinth of branching tubules and flattened sacs
the rough ER has many ribosomes bound to its cytosolic surface, and is a major site or protein production
proteins are transported into the ER as they are synthesized
the ER membrane is the site of production of all the transmembrane proteins
also produces most of the lipid for the rest of the cell
almost all secreted proteins to the cell exterior are initially delivered to the ER lumen
stores Ca2+ ions
in liver cells, the ER has a surface area 25x that of the plasma membrane
Smooth ER
the transitional ER is a smooth ER from which transport vesicles are budding off
in cells that specialize in lipid metabolism, the smooth ER is abundant
the ER store enzymes (cytochrome P450 family) that metabolize lipid-soluble drugs (rendered water soluble to leave the cell and be excreted in the urine)
Nuclear Envelope/ER Figure
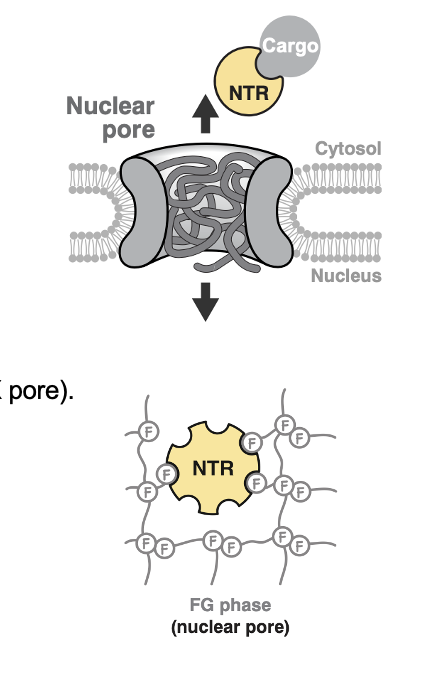
Nuclear Import
nuclear import requires NLS + importin (nuclear transport receptor) + Ran-GTP cycle (energy) to get through
Nuclear pore complexes perforate the nuclear envelope
some proteins continually shuttle back and forth between the nucleus and the cytosol
the relative rates of their import and export determine the steady-state localization
Nuclear Pore Complex (NPC): Structure
selective hydrogel-like barrier formed by FG repeats (Phe-Glyc)
3000-4000 NPCs per cell
built from ~30 different proteins (nucleoporins)
unlike PEX pores, the NPC isn’t a simple membrane channel, it sits outside the bilayer like a giant ring complex
NPCs: FG-Fibrils
the pore is gated by FG-fibrils
FG= Phe-Gly repeats, which form unstructured, hydrophobic tendrils inside the pore (a hydrogel-like mesh)
the mesh blocks passive diffusion of large molecules (>60 kDa)
ribosomes are ~30nm in diameter and thus cannot diffuse thru the NPC
only proteins with the right importins can get through
Nuclear transporter receptors (NTRs) diffuse thru the FG meshwork by transiently binding to F residues using hydrophobic patches
NPCs: How transport works
importins have hydrophobic patches and are specialized for the transport of a subset of cargo proteins
importins bind the NLS and bind Phe in FG repeats transiently
hop from FG-repeat to FG-repeat
this allows selective passage thru the mesh
NLS
nuclear localization signals are responsible for the nuclear import process
short Lys-Arg rich positive sequence
signal is located anywhere in the protein (loops or patches on protein surface), the precise location of the signal within the protein is not important
fully folded proteins can be transported into the nucleus thru an NPC
NPC Figure
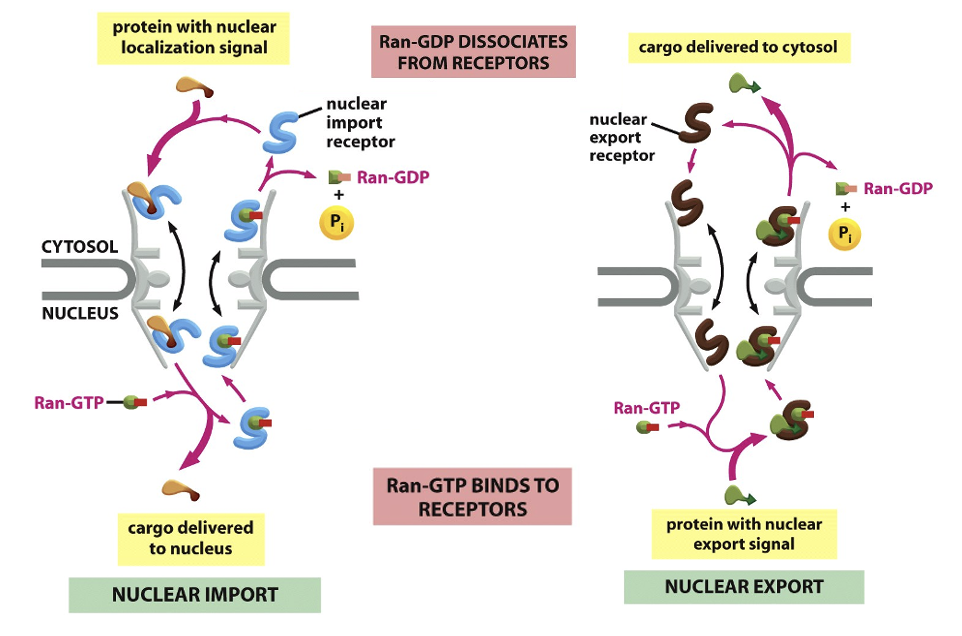
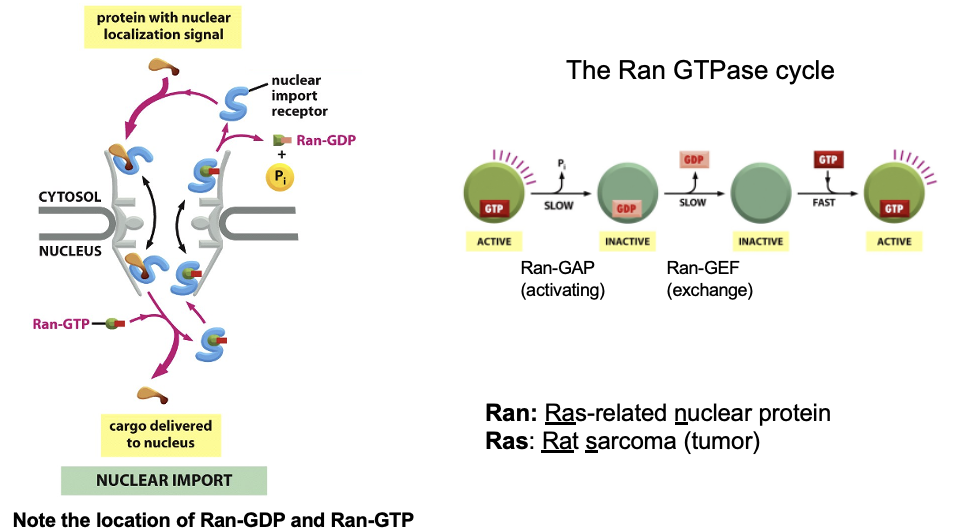
Nuclear Import Transport model: Import
cargo binds importin in the cytosol (importins recognize NLS sequences)
importin-cargo moves thru the FG-mesh of the NPC
Ran-GTP binds to the importin on the nuclear side of the pore
Ran-GTP binding causes the receptors to release their cargo
the importin with Ran-GTP is transported back to the cytosol
Ran-GAP in the cytosol triggers the hydrolysis of GTP, thereby converting it to Ran-GDP
the import receptor is then ready for another cycle of nuclear import
Nuclear Import Transport model: Export
exportins binds both cargo and Ran-GTP inside the nucleus (export cargos typically have nuclear export signals)
exportin-cargo-Ran-GTP passes thru the pore
in the cytosol, Ran-GTP hydrolyzes GTP → GDP
the export-receptor releases both its cargo and Ran-GDP in the cytosol
free export receptors are then returned to the nucleus to complete the cycle
Nuclear Import Transport: Gradient
the import cycle is powered by a Ran-GTP gradient across the pore
the gradient arises because of the exclusive nuclear localiztion of proteins called Ran GEFs
Ran-GEF is only in the nucleus, GEF=guanine exchange factor → loads GTP onto Ran to produce Ran-GTP
these proteins exchange GDP for GTP on Ran molecules
there is an elevated RanGTP concetration in the nucleus compared to the cytoplasm
Ran-GAP is only in the cytosol
GAP= GTPase activating protein → hydrolyzes Ran-GTP → Ran GDP
Nuclear Import Transport Model Figure
Ran Cycling Mechanism
translocation thru the pore is not energy-dependent; the cargo passes thru the pore with the assistance of importins
the whole import cycle and directionality requires Ran-GTP hydrolysis (but not used energy to push cargo physically, rather it resets the receptors)
the driving force for transport depends on the gradient of Ran-GTP
the gradient of Ran-GTP is made because of the preferential location of Ran-DEG and Ran-GAP
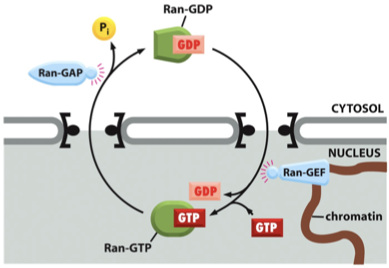
Ran Cycling Mechanism: Regulatory Proteins
Ran-GEF (nuclear) converts Ran-GDP → Ran-GTP
a GTP exchange factor
Ran-GAP (cytosolic) converts Ran-GTP→Ran-GDP
a GTPase-activating protein
Ran Cycling Mechanism:
The Piggy-Back Transport Model
the cargo itself doesn’t need a signal if it associates with signal-bearing partner